1TIB
 
 | |
5GTW
 
 | |
1GG6
 
 | | CRYSTAL STRUCTURE OF GAMMA CHYMOTRYPSIN WITH N-ACETYL-PHENYLALANINE TRIFLUOROMETHYL KETONE BOUND AT THE ACTIVE SITE | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), 1,2-ETHANEDIOL, GAMMA CHYMOTRYPSIN, ... | | Authors: | Neidhart, D, Wei, Y, Cassidy, C, Lin, J, Cleland, W.W, Frey, P.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin.
Biochemistry, 40, 2001
|
|
1TIA
 
 | | AN UNUSUAL BURIED POLAR CLUSTER IN A FAMILY OF FUNGAL LIPASES | | Descriptor: | LIPASE | | Authors: | Derewenda, U, Swenson, L, Yamaguchi, S, Wei, Y, Derewenda, Z.S. | | Deposit date: | 1993-12-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An unusual buried polar cluster in a family of fungal lipases.
Nat.Struct.Biol., 1, 1994
|
|
4LX3
 
 | | Conserved Residues that Modulate Protein trans-Splicing of Npu DnaE Split Intein | | Descriptor: | DNA polymerase III, alpha subunit, Nucleic acid binding, ... | | Authors: | Wu, Q, Gao, Z, Wei, Y, Ma, G, Zheng, Y, Dong, Y, Liu, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved residues that modulate protein trans-splicing of Npu DnaE split intein.
Biochem.J., 461, 2014
|
|
5GOL
 
 | |
6BY9
 
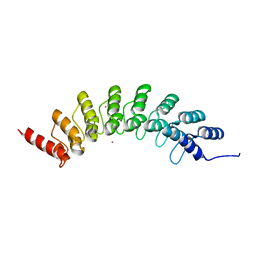 | | Crystal structure of EHMT1 | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Wei, Y, Li, A, Tempel, W, Han, S, Sunnerhagen, M, Penn, L, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of EHMT1
to be published
|
|
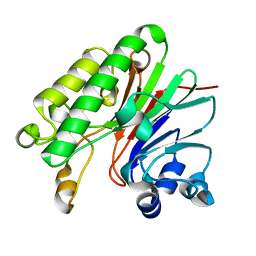
4F1R
 
 | |
6LM3
 
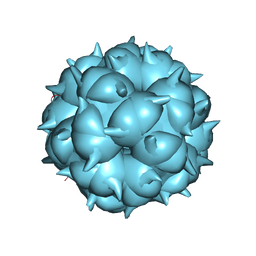 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
4XO2
 
 | |
1JXZ
 
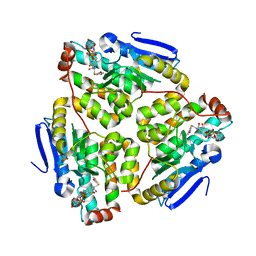 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
7LQT
 
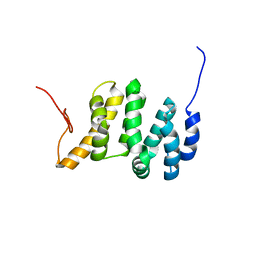 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
4EEQ
 
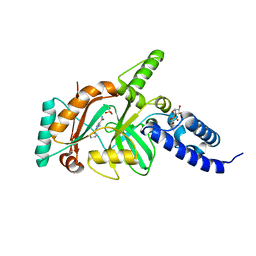 | | Crystal structure of E. faecalis DNA ligase with inhibitor | | Descriptor: | 4-amino-2-(cyclopentyloxy)pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wang, T, Charifson, P, Wei, Y. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis and Activity Evaluation of Potent NAD+ DNA Ligase Inhibitors as Potential Antibacterial Agents.
To be Published
|
|
1TLH
 
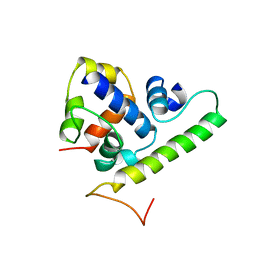 | | T4 AsiA bound to sigma70 region 4 | | Descriptor: | 10 kDa anti-sigma factor, RNA polymerase sigma factor rpoD | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma(70) region 4
Embo J., 23, 2004
|
|
1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1TL6
 
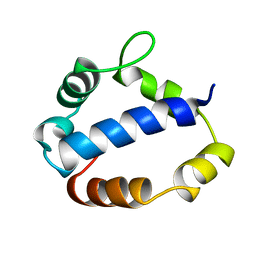 | | Solution structure of T4 bacteriphage AsiA monomer | | Descriptor: | 10 kDa anti-sigma factor | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma70 region 4
Embo J., 23, 2004
|
|
1TTY
 
 | | Solution structure of sigma A region 4 from Thermotoga maritima | | Descriptor: | RNA polymerase sigma factor rpoD | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-23 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma(70) region 4
Embo J., 23, 2004
|
|
3TKA
 
 | | crystal structure and solution saxs of methyltransferase rsmh from E.coli | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Ribosomal RNA small subunit methyltransferase H, S-ADENOSYLMETHIONINE, ... | | Authors: | Gao, Z.Q, Wei, Y, Zhang, H, Dong, Y.H. | | Deposit date: | 2011-08-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA.
J.Struct.Biol., 179, 2012
|
|
2GF5
 
 | | Structure of intact FADD (MORT1) | | Descriptor: | FADD protein | | Authors: | Carrington, P.E, Sandu, C, Wei, Y, Hill, J.M, Morisawa, G, Huang, T, Gavathiotis, E, Wei, Y, Werner, M.H. | | Deposit date: | 2006-03-21 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of FADD and Its Mode of Interaction with Procaspase-8
Mol.Cell, 22, 2006
|
|
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | Descriptor: | Glycoprotein, scFv 523-11 | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9037 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | Descriptor: | Glycoprotein,Glycoprotein,Glycoprotein | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
5X8I
 
 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
6CVZ
 
 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6JHB
 
 | |
