4N47
 
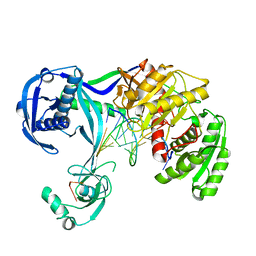 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 12-mer target DNA | | 分子名称: | 5'-D(*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-08 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.823 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4XZE
 
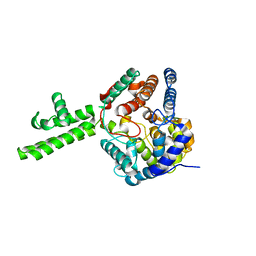 | | The crystal structure of Hazara virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZA
 
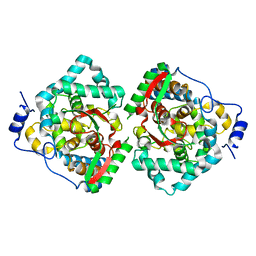 | | The crystal structure of Erve virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
2FXZ
 
 | |
2FXY
 
 | |
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | 著者 | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | 登録日 | 2002-08-23 | | 公開日 | 2002-12-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.52 Å) | | 主引用文献 | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
8W8Q
 
 | | Cryo-EM structure of the GPR101-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8R
 
 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | 分子名称: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | 分子名称: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | 著者 | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
1W63
 
 | | AP1 clathrin adaptor core | | 分子名称: | ADAPTER-RELATED PROTEIN COMPLEX 1 BETA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 SIGMA 1A SUBUNIT, ... | | 著者 | Heldwein, E, Macia, E, Wang, J, Yin, H.L, Kirchhausen, T, Harrison, S.C. | | 登録日 | 2004-08-12 | | 公開日 | 2004-09-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Crystal Structure of the Clathrin Adaptor Protein 1 Core
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2GF9
 
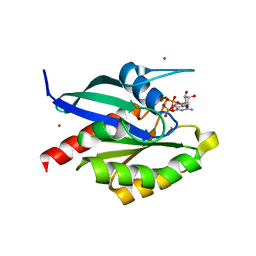 | | Crystal structure of human RAB3D in complex with GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-3D, ... | | 著者 | Hong, B, Wang, J, Shen, L, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2006-03-21 | | 公開日 | 2006-05-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Crystal structure of human RAB3D in complex with GDP
To be Published
|
|
7YOJ
 
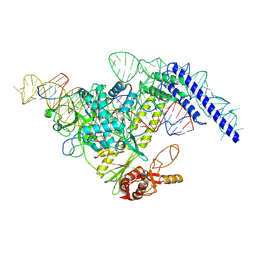 | | Structure of CasPi with guide RNA and target DNA | | 分子名称: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | 著者 | Li, C.P, Wang, J, Liu, J.J. | | 登録日 | 2022-08-01 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-06 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | 分子名称: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-05 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
5XKU
 
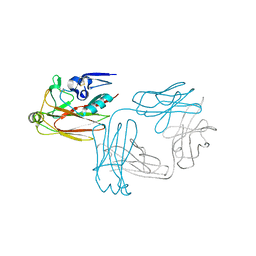 | | Crystal structure of hemagglutinin globular head from an H7N9 influenza virus in complex with a neutralizing antibody HNIgGA6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HNIgGA6 heavy chain, HNIgGA6 light chain, ... | | 著者 | Chen, C, Wang, J, Wang, W, Gao, X, Cui, S, Jin, Q. | | 登録日 | 2017-05-09 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural Insight into a Human Neutralizing Antibody against Influenza Virus H7N9
J. Virol., 92, 2018
|
|
3U83
 
 | | Crystal structure of nectin-1 | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Poliovirus receptor-related protein 1 | | 著者 | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2011-10-15 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.499 Å) | | 主引用文献 | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
3U82
 
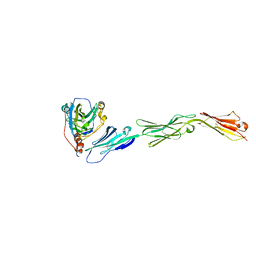 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | 分子名称: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | 著者 | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2011-10-15 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.164 Å) | | 主引用文献 | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
2QD0
 
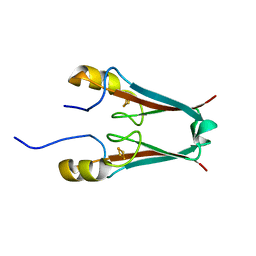 | | Crystal structure of mitoNEET | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | 著者 | Lin, J, Zhou, T, Ye, K, Wang, J. | | 登録日 | 2007-06-20 | | 公開日 | 2007-08-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1MHK
 
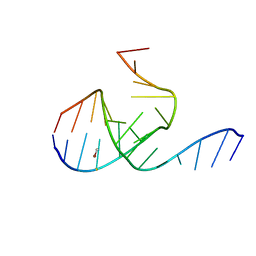 | | Crystal Structure Analysis of a 26mer RNA molecule, representing a new RNA motif, the hook-turn | | 分子名称: | BROMIDE ION, RNA 12-mer BCh12, RNA 14-mer BCh12 | | 著者 | Szep, S, Wang, J, Moore, P.B. | | 登録日 | 2002-08-20 | | 公開日 | 2002-09-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of a 26-nucleotide RNA containing a hook-turn
RNA, 9, 2003
|
|
4MSB
 
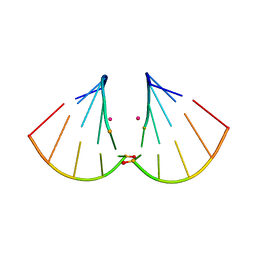 | | RNA 10mer duplex with two 2'-5'-linkages | | 分子名称: | RNA 10mer duplex with two 2'-5'-linkages, STRONTIUM ION | | 著者 | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | 登録日 | 2013-09-18 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | 分子名称: | 3C-like proteinase, GLYCEROL, UAW243 | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-15 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | 分子名称: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-06 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | 分子名称: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
4XSJ
 
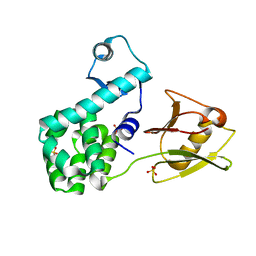 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | 分子名称: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | 著者 | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | 登録日 | 2015-01-22 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4MS9
 
 | | Native RNA-10mer Structure: ccggcgccgg | | 分子名称: | Native RNA duplex 10mer, STRONTIUM ION | | 著者 | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | 登録日 | 2013-09-18 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
