5X7U
 
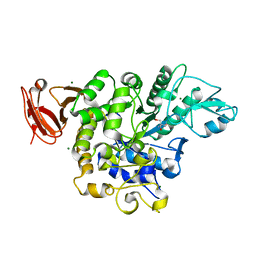 | | Trehalose synthase from Thermobaculum terrenum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Trehalose synthase | | Authors: | Su, J, Wang, F. | | Deposit date: | 2017-02-27 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Characteristics and Function of a New Kind of Thermostable Trehalose Synthase from Thermobaculum terrenum.
J. Agric. Food Chem., 65, 2017
|
|
3RLQ
 
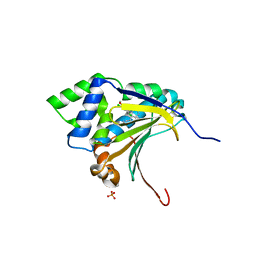 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5- carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLR
 
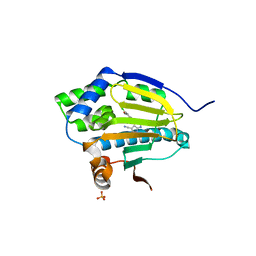 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6U8B
 
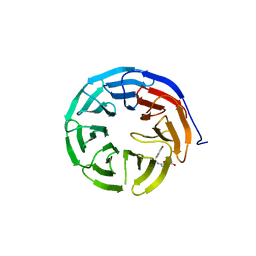 | |
6U8L
 
 | |
3RLP
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
8V1P
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092 | | Descriptor: | Glucose-induced degradation protein 4 homolog, N,N~2~-bis[(4-methoxyphenyl)methyl]glycinamide | | Authors: | Dong, C, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092
To be published
|
|
6U8O
 
 | |
5XHX
 
 | |
5EPO
 
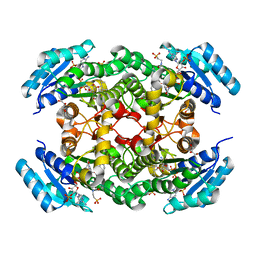 | | The three-dimensional structure of Clostridium absonum 7alpha-hydroxysteroid dehydrogenase | | Descriptor: | 7-alpha-hydroxysteroid deydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lou, D, Wang, B, Wang, F. | | Deposit date: | 2015-11-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Clostridium absonum 7 alpha-hydroxysteroid dehydrogenase: new insights into the conserved arginines for NADP(H) recognition
Sci Rep, 6, 2016
|
|
8FK0
 
 | |
1HM1
 
 | | THE SOLUTION NMR STRUCTURE OF A THERMALLY STABLE FAPY ADDUCT OF AFLATOXIN B1 IN AN OLIGODEOXYNUCLEOTIDE DUPLEX REFINED FROM DISTANCE RESTRAINED MOLECULAR DYNAMICS SIMULATED ANNEALING, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*TP*AP*TP*(FAG)P*AP*TP*TP*CP*A)-3'), DNA (5'-D(TP*GP*AP*AP*TP*CP*AP*TP*AP*G)-3') | | Authors: | Mao, H, Deng, Z, Wang, F, Harris, T.M, Stone, M.P. | | Deposit date: | 1998-05-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR.
Biochemistry, 37, 1998
|
|
2GIZ
 
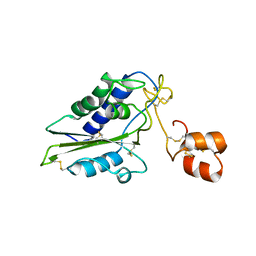 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
6K9N
 
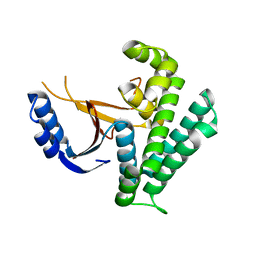 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6U80
 
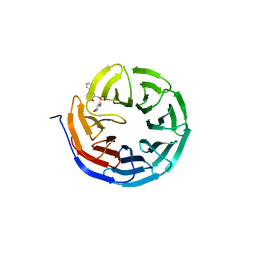 | |
6U7M
 
 | |
6U5Y
 
 | |
6U5M
 
 | |
8EWG
 
 | | Cryo-EM structure of a riboendonclease | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | Authors: | Li, Z, Wang, F. | | Deposit date: | 2022-10-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
8FJS
 
 | |
3JCI
 
 | | 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles | | Descriptor: | Capsid protein | | Authors: | Liu, Z, Guo, F, Wang, F, Li, T.C, Jiang, W. | | Deposit date: | 2015-12-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | 2.9 angstrom Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles.
Structure, 24, 2016
|
|
4Y2I
 
 | | Gold ion bound to GolB | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
4Y2K
 
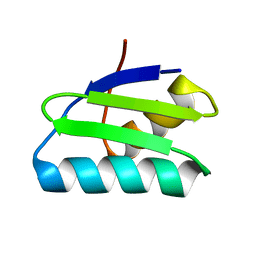 | | reduced form of apo-GolB | | Descriptor: | Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
4Y2M
 
 | | apo-GolB protein | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Zhao, J, Wang, F. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
