4Z31
 
 | | Crystal structure of the RC3H2 ROQ domain in complex with stem-loop and double-stranded forms of RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*A)-D(P*UP*GP*UP*UP*CP*UP*GP*UP*GP*AP*AP*CP*AP*C)-3'), Roquin-2, ... | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins.
Sci Rep, 5, 2015
|
|
1W5C
 
 | | Photosystem II from Thermosynechococcus elongatus | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Biesiadka, J, Loll, B, Kern, J, Irrgang, K.-D, Saenger, W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-21 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Cyanobacterial Photosystem II at 3.2 A Resolution: A Closer Look at the Mn- Cluster
Phys.Chem.Chem.Phys., 6, 2004
|
|
2WF9
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, and Beryllium trifluoride, crystal form 2 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WF8
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, Glucose-1-phosphate and Beryllium trifluoride | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Near attack conformers dominate beta-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
2WA0
 
 | | Crystal structure of the human MAGEA4 | | Descriptor: | MELANOMA-ASSOCIATED ANTIGEN 4 | | Authors: | Roos, A.K, Cooper, C.D.O, Ugochukwu, E, W Yue, W, Berridge, G, Elkins, J.M, Pike, A.C.W, Bray, J, Filippakopoulos, P, Muniz, J, Chaikuad, A, Burgess-Brown, N, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O, Oppermann, U. | | Deposit date: | 2009-01-31 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding.
Plos One, 11, 2016
|
|
2WFA
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WF6
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and Aluminium tetrafluoride | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2WF7
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphonate and Aluminium tetrafluoride | | Descriptor: | 6,7-dideoxy-7-phosphono-beta-D-gluco-heptopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Alpha-Fluorophosphonates Reveal How a Phosphomutase Conserves Transition State Conformation Over Hexose Recognition in its Two-Step Reaction.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5AC7
 
 | | S. enterica HisA with mutations D7N, D10G, dup13-15 | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SODIUM ION, ... | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7VMU
 
 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | Spike protein S1, scFv E4 | | Authors: | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
5AC8
 
 | | S. enterica HisA with mutations D10G, dup13-15, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2RUR
 
 | | Solution structure of Human Pin1 PPIase C113S mutant | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
4Y72
 
 | | Human CDK1/CyclinB1/CKS2 With Inhibitor | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
4YAB
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 1-methyl-5-(2-methyl-1 3-thiazol-4-yl)-2 3-dihydro-1H-indol-2-one (1) | | Descriptor: | 1-methyl-5-(2-methyl-1,3-thiazol-4-yl)-1,3-dihydro-2H-indol-2-one, SULFATE ION, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAD
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 2,4-dimethoxy-N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzene-1-sulfonamide (3b) | | Descriptor: | 2,4-dimethoxy-N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzenesulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YC3
 
 | | CDK1/CyclinB1/CKS2 Apo | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-19 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
5AB3
 
 | | S.enterica HisA mutant D7N, D10G, dup13-15, Q24L, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SODIUM ION, [(2R,3S,4R,5R)-5-[4-AMINOCARBONYL-5-[[(Z)-[(3R,4R)-3,4-DIHYDROXY-2-OXO-5-PHOSPHONOOXY-PENTYL]IMINOMETHYL]AMINO]IMIDAZOL-1-YL]-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5AK7
 
 | | Structure of wt Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
4YBM
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide (7b) | | Descriptor: | GLYCEROL, N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-3,4-dimethoxybenzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBS
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide (7g) | | Descriptor: | DIMETHYL SULFOXIDE, N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
5AJR
 
 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma cruzi in complex with the 1-tetrazole derivative VT-1161 ((R)-2-(2,4-Difluorophenyl)-1, 1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl) pyridin-2-yl)propan-2-ol) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Wawrzak, Z, Hoekstra, W, I Lepesheva, G. | | Deposit date: | 2015-02-26 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Clinical Candidate Vt-1161'S Antiparasitic Effect in Vitro, Activity in a Murine Model of Chagas Disease, and Structural Characterization in Complex with the Target Enzyme Cyp51 from Trypanosoma Cruzi.
Antimicrob.Agents Chemother., 60, 2015
|
|
1YDO
 
 | | Crystal Structure of the Bacillis subtilis HMG-CoA Lyase, Northeast Structural Genomics Target SR181. | | Descriptor: | HMG-CoA Lyase, IODIDE ION | | Authors: | Forouhar, F, Hussain, M, Edstrom, W, Vorobiev, S.M, Xiao, R, Ciano, M, Shih, L, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.
J.Biol.Chem., 281, 2006
|
|
4XEM
 
 | |
5AK8
 
 | | Structure of C351A mutant of Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
2WNJ
 
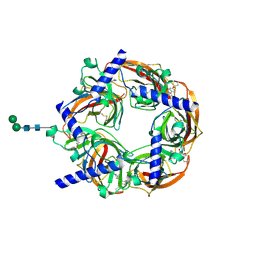 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH DMXBA | | Descriptor: | (3E)-3-[(2,4-DIMETHOXYPHENYL)METHYLIDENE]-3,4,5,6-TETRAHYDRO-2,3'-BIPYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants for Interaction of Partial Agonists with Acetylcholine Binding Protein and Neuronal Alpha7 Nicotinic Acetylcholine Receptor.
Embo J., 28, 2009
|
|
