1IEH
 
 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
1J1N
 
 | | Structure Analysis of AlgQ2, A Macromolecule(Alginate)-Binding Periplasmic Protein Of Sphingomonas Sp. A1., Complexed with an Alginate Tetrasaccharide | | Descriptor: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mikami, B, Mishima, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2002-12-11 | | Release date: | 2003-06-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-A resolution
J.BIOL.CHEM., 278, 2003
|
|
1IJ5
 
 | | METAL-FREE STRUCTURE OF MULTIDOMAIN EF-HAND PROTEIN, CBP40, FROM TRUE SLIME MOLD | | Descriptor: | PLASMODIAL SPECIFIC LAV1-2 PROTEIN | | Authors: | Iwasaki, W, Sasaki, H, Nakamura, A, Kohama, K, Tanokura, M. | | Deposit date: | 2001-04-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Metal-Free and Ca(2+)-Bound Structures of a Multidomain EF-Hand Protein, CBP40, from the Lower Eukaryote Physarum polycephalum
Structure, 11, 2003
|
|
6NU4
 
 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
3LO9
 
 | |
4H2M
 
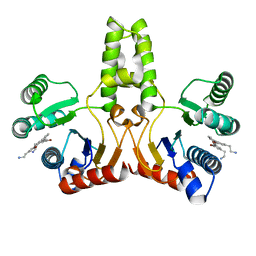 | | Structure of E. coli undecaprenyl diphosphate synthase in complex with BPH-1408 | | Descriptor: | 2,2'-{benzene-1,3-diylbis[ethyne-2,1-diyl(5-bromobenzene-3,1-diyl)]}diethanamine, Undecaprenyl pyrophosphate synthase | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2012-09-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Antibacterial drug leads targeting isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3LVX
 
 | |
3LXH
 
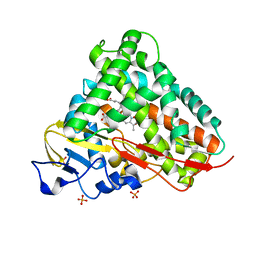 | | Crystal Structure of Cytochrome P450 CYP101D1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LU1
 
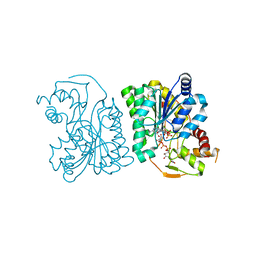 | | Crystal Structure Analysis of WbgU: a UDP-GalNAc 4-epimerase | | Descriptor: | GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bhatt, V.S, Guo, C.Y, Zhao, G, Yi, W, Liu, Z.J, Wang, P.G. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases.
Protein Sci., 20, 2011
|
|
4H3A
 
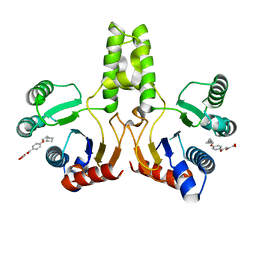 | |
3LXD
 
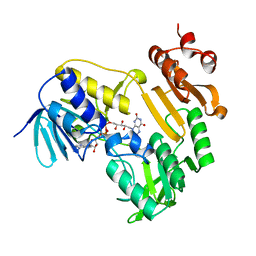 | | Crystal Structure of Ferredoxin Reductase ArR from Novosphingobium aromaticivorans | | Descriptor: | FAD-dependent pyridine nucleotide-disulphide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
4H2O
 
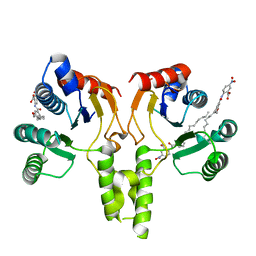 | |
3CNI
 
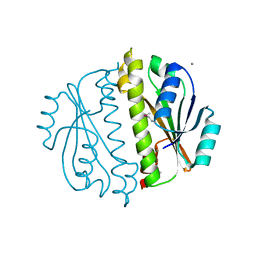 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
3CRM
 
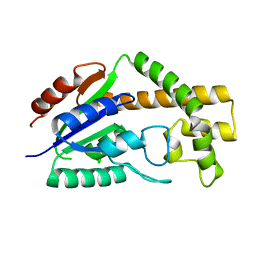 | |
3CON
 
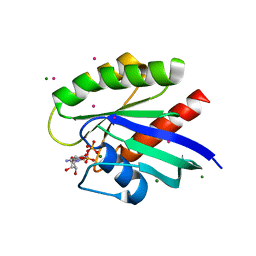 | | Crystal structure of the human NRAS GTPase bound with GDP | | Descriptor: | GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Shen, L, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal structure of the human NRAS GTPase bound with GDP.
To be Published
|
|
3COS
 
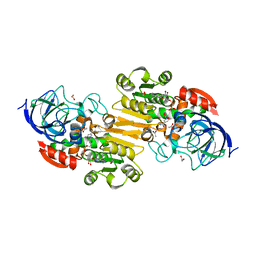 | | Crystal structure of human class II alcohol dehydrogenase (ADH4) in complex with NAD and Zn | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Alcohol dehydrogenase 4, ... | | Authors: | Kavanagh, K.L, Shafqat, N, Yue, W, von Delft, F, Bishop, S, Roos, A, Murray, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human class II alcohol dehydrogenase (ADH4) in complex with NAD and Zn.
To be Published
|
|
1WBY
 
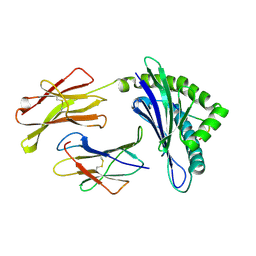 | | CRYSTAL STRUCTURES OF MURINE MHC CLASS I H-2 Db AND Kb MOLECULES IN COMPLEX WITH CTL EPITOPES FROM INFLUENZA A VIRUS: IMPLICATIONS FOR TCR REPERTOIRE SELECTION AND IMMUNODOMINANCE | | Descriptor: | BETA-2MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Meijers, R, Lai, C, Yang, Y, Liu, J, Zhong, W, Wang, J, Reinherz, E.L. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Murine Mhc Class I H-2 D(B) and K(B) Molecules in Complex with Ctl Epitopes from Influenza a Virus: Implications for Tcr Repertoire Selection and Immunodominance
J.Mol.Biol., 345, 2005
|
|
1X55
 
 | | Crystal structure of asparaginyl-tRNA synthetase from Pyrococcus horikoshii complexed with asparaginyl-adenylate analogue | | Descriptor: | 5'-O-[N-(L-ASPARAGINYL)SULFAMOYL]ADENOSINE, Asparaginyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Water-assisted Asparagine Recognition by Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 360, 2006
|
|
3D81
 
 | | Sir2-S-alkylamidate complex crystal structure | | Descriptor: | NAD-dependent deacetylase, S-alkylamidate intermediate, ZINC ION | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
6ZBV
 
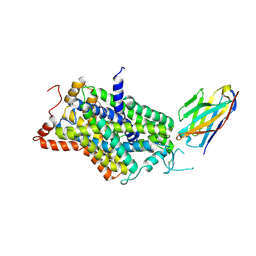 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor and sybody Sb_GlyT1#7 | | Descriptor: | Sodium- and chloride-dependent glycine transporter 1,Sodium- and chloride-dependent glycine transporter 1, Sybody Sb_GlyT1#7, [5-fluoranyl-6-(oxan-4-yloxy)-1,3-dihydroisoindol-2-yl]-[5-methylsulfonyl-2-[2,2,3,3,3-pentakis(fluoranyl)propoxy]phenyl]methanone | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
1X1J
 
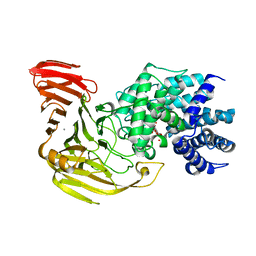 | | Crystal Structure of Xanthan Lyase (N194A) with a Substrate. | | Descriptor: | (4AR,6R,7S,8R,8AR)-8-((5R,6R)-3-CARBOXY-TETRAHYDRO-4,5,6-TRIHYDROXY-2H-PYRAN-2-YLOXY)-HEXAHYDRO-6,7-DIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID), CALCIUM ION, xanthan lyase | | Authors: | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-04-04 | | Release date: | 2005-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
3CP8
 
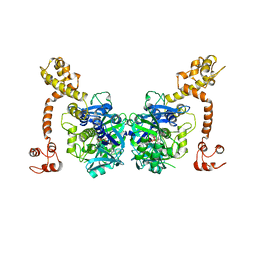 | | Crystal structure of GidA from Chlorobium tepidum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Meyer, S, Scrima, A, Versees, W, Wittinghofer, A. | | Deposit date: | 2008-03-31 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
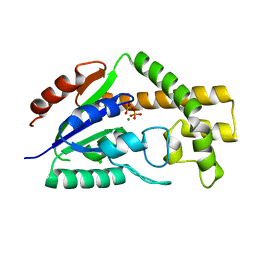
3CRQ
 
 | |
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
3CZW
 
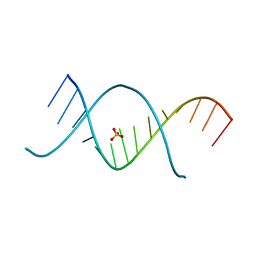 | | X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex | | Descriptor: | RNA (5'-R(*G*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3'), SULFATE ION | | Authors: | Rypniewski, W, Adamiak, D.A, Milecki, J, Adamiak, R.W. | | Deposit date: | 2008-04-30 | | Release date: | 2008-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Noncanonical G(syn)-G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex.
Rna, 14, 2008
|
|
