6PKJ
 
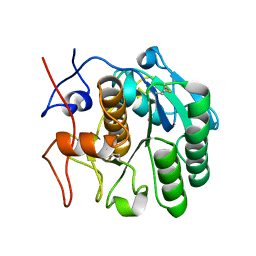 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#2) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKO
 
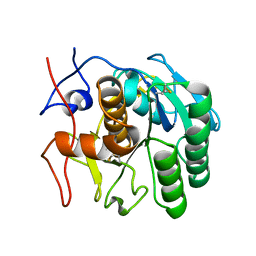 | | MicroED structure of proteinase K from a platinum coated, unpolished, single lamella at 2.07A resolution (#12) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.07 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKN
 
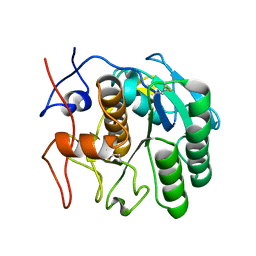 | | MicroED structure of proteinase K from an unpolished, platinum-coated, single lamella at 2.08A resolution (#9) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.08 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6VCH
 
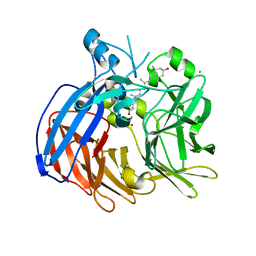 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase in complex with 3-hydroxy-beta-apo-14'-carotenal | | Descriptor: | (2E,4E,6E,8E,10E)-11-[(4R)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-5,9-dimethylundeca-2,4,6,8,10-pentaenal, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VCC
 
 | | Cryo-EM structure of the Dvl2 DIX filament | | Descriptor: | Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Enos, M, Kan, W, Muennich, S, Chen, D.H, Skiniotis, G, Weis, W.I. | | Deposit date: | 2019-12-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Limited Dishevelled/Axin oligomerization determines efficiency of Wnt/ beta-catenin signal transduction.
Elife, 9, 2020
|
|
6VCN
 
 | | Crystal structure of E.coli RppH in complex with ppcpG | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCS
 
 | | SRA domain of UHRF1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*AP*CP*AP*GP*GP*C)-3'), E3 ubiquitin-protein ligase UHRF1, UNK-UNK-UNK-UNK, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SRA domain of UHRF1 in complex with DNA
To Be Published
|
|
6PKR
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.79A resolution (#13) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.79 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6V3N
 
 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6VAU
 
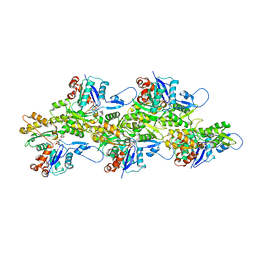 | | Bare actin filament from a partially cofilin-decorated sample | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1SGN
 
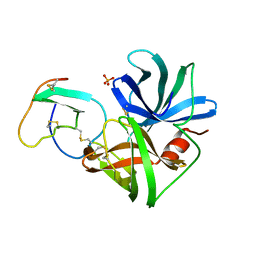 | | ASN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with
Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1SGY
 
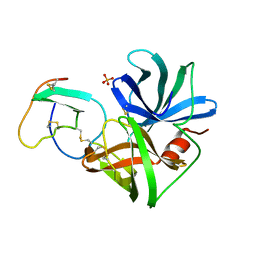 | | TYR 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
6L5A
 
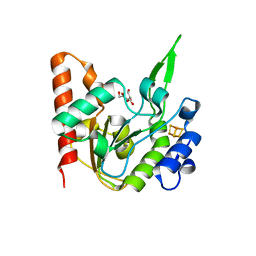 | | The structure of the UdgX mutant H109E at a pre-excision state | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Xie, W, Tu, J, Zeng, H. | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.80007327 Å) | | Cite: | Structural insights into an MsmUdgX mutant capable of both crosslinking and uracil excision capability.
DNA Repair (Amst), 97, 2021
|
|
6PKK
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.18A resolution (#5) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKP
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.91A resolution (#10) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
1NPC
 
 | |
6VEC
 
 | | Cryo-EM structure of F-actin/Plastin2-ABD2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Zheng, W, Kudryashov, D.S, Egelman, E.H. | | Deposit date: | 2019-12-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Osteogenesis imperfecta mutations in plastin 3 lead to impaired calcium regulation of actin bundling.
Bone Res, 8, 2020
|
|
1L7M
 
 | | HIGH RESOLUTION LIGANDED STRUCTURE OF PHOSPHOSERINE PHOSPHATASE (PI COMPLEX) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6PQ0
 
 | | LCP-embedded Proteinase K treated with MPD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
4IQB
 
 | | High Resolution Crystal Structure of C.elegans Thymidylate Synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
6POM
 
 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6VAO
 
 | | Human cofilin-1 decorated actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1FZW
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). APO ENZYME. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-04 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
