3KBH
 
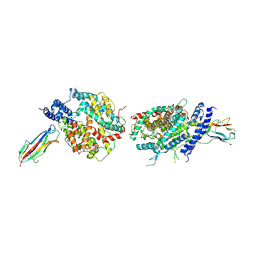 | | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Wu, K, Li, W, Peng, G, Li, F. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6BSG
 
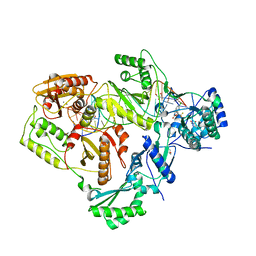 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in an RNA hydrolysis-off mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3KEF
 
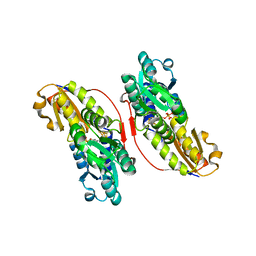 | | Crystal structure of IspH:DMAPP-complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, DIMETHYLALLYL DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5UHG
 
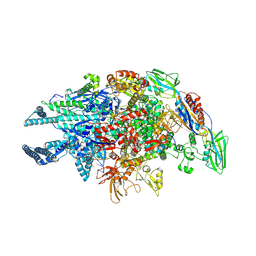 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with D-AAP1 and Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.971 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
4PDV
 
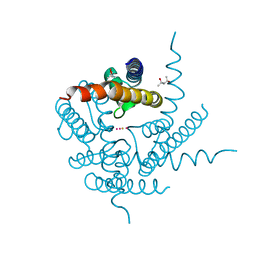 | | Structure of K+ selective NaK mutant in barium and potassium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BARIUM ION, POTASSIUM ION, ... | | Authors: | Lam, Y, Zeng, W, Sauer, D.B, Jiang, Y. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | High Resolution Structural Views of Rubidium, Cesium and Barium Binding within a Potassium Selective Channel Filter
To Be Published
|
|
6C6K
 
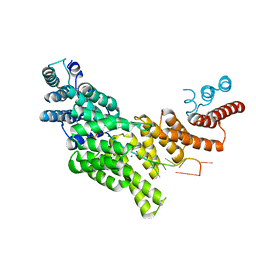 | | Structural basis for preferential recognition of cap 0 RNA by a human IFIT1-IFIT3 protein complex | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, Interferon-induced protein with tetratricopeptide repeats 3, MAGNESIUM ION, ... | | Authors: | Amarasinghe, G.K, Leung, D.W, Johnson, B, Xu, W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability.
Immunity, 48, 2018
|
|
3HPC
 
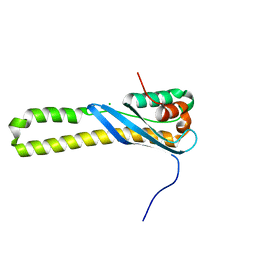 | |
2NS2
 
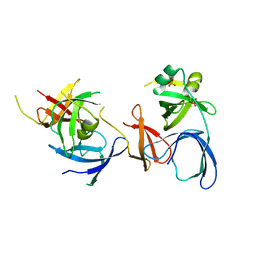 | | Crystal Structure of Spindlin1 | | Descriptor: | PHOSPHATE ION, Spindlin-1 | | Authors: | Zhao, Q, Qin, L, Jiang, F, Wu, B, Yue, W, Xu, F, Rong, Z, Yuan, H, Xie, X, Gao, Y, Bai, C, Bartlam, M. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation
J.Biol.Chem., 282, 2007
|
|
3HXE
 
 | | Engineered RabGGTase in complex with a peptidomimetic inhibitor (compound 37) | | Descriptor: | CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, Geranylgeranyl transferase type-2 subunit beta, ... | | Authors: | Guo, Z, Alexandrov, K, Waldmann, H, Goody, R.S, Blankenfeldt, W. | | Deposit date: | 2009-06-20 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, and characterization of Peptide-based rab geranylgeranyl transferase inhibitors
J.Med.Chem., 52, 2009
|
|
6IIL
 
 | | USP14 catalytic domain bind to IU1-47 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(piperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, Wang, F, Wang, J.W, He, W, Ding, S, Li, J.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
7GHJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with SIM-SYN-f15aaa3a-1 (Mpro-x3348) | | Descriptor: | 1-[4-(diphenylmethyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2NTA
 
 | | Crystal Structure of PTP1B-inhibitor Complex | | Descriptor: | 5-(4-CHLORO-5-PHENYL-3-THIENYL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Follows, B. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing acid replacements of thiophene PTP1B inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7GHK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAR-DIA-eace69ff-36 (Mpro-x3351) | | Descriptor: | 1-(5-fluoro-1H-indol-3-yl)-N-methylmethanamine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4QQI
 
 | | Crystal structure of ANKRA2-RFX7 complex | | Descriptor: | Ankyrin repeat family A protein 2, DNA-binding protein RFX7, SULFATE ION, ... | | Authors: | Xu, C, Tempel, W, Dong, A, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of ANKRA2-RFX7 complex
To Be Published
|
|
3HVQ
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Neurabin | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Neurabin-1, ... | | Authors: | Critton, D.A, Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2NVV
 
 | | Crystal Structure of the Putative Acetyl-CoA hydrolase/transferase PG1013 from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR16. | | Descriptor: | Acetyl-CoA hydrolase/transferase family protein, ZINC ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Yong, W, Ho, C.K, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Acetyl-CoA hydrolase/transferase PG1013 from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR16
To be Published
|
|
4QUY
 
 | | yCP beta5-A49S-mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3I2E
 
 | | Crystal structure of human dimethylarginine dymethylaminohydrolase-1 (DDAH-1) | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Monzingo, A.F, Wang, Y, Hu, S, Schaller, T.H, Robertus, J.D, Fast, W. | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Developing dual and specific inhibitors of dimethylarginine dimethylaminohydrolase-1 and nitric oxide synthase: toward a targeted polypharmacology to control nitric oxide.
Biochemistry, 48, 2009
|
|
2NXW
 
 | | Crystal structure of phenylpyruvate decarboxylase of Azospirillum brasilense | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Versees, W, Spaepen, S, Vanderleyden, J, Steyaert, J. | | Deposit date: | 2006-11-20 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 A resolution. Implications for its catalytic and regulatory mechanism.
Febs J., 274, 2007
|
|
4QW3
 
 | | yCP beta5-C63F mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3HXG
 
 | | Crystal structure of Schistsome eIF4E complexed with m7GpppA and 4E-BP | | Descriptor: | Eukaryotic Translation Initiation Factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Liu, W, Zhao, R, Jones, D.N.M, Davis, R.E. | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
J.Biol.Chem., 284, 2009
|
|
6AOZ
 
 | |
4QYY
 
 | | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase State | | Descriptor: | (3R)-1-{2-[4-(4-acetylphenyl)piperazin-1-yl]-2-oxoethyl}-N-(3-chloro-4-hydroxyphenyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Deng, Y, Shipps, G.W, Cooper, A, English, J.M, Annis, D.A, Carr, D, Nan, Y, Wang, T, Zhu, Y.H, Chuang, C, Dayananth, P, Hruza, A.W, Xiao, L, Jin, W, Kirschmeier, P, Windsor, W.T, Samatar, A.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase.
J.Med.Chem., 57, 2014
|
|
4QPL
 
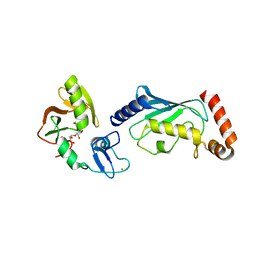 | | Crystal structure of RNF146(RING-WWE)/UbcH5a/iso-ADPr complex | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Wang, Z, DaRosa, P.A, Klevit, R.E, Xu, W. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal.
Nature, 517, 2015
|
|
4QZ3
 
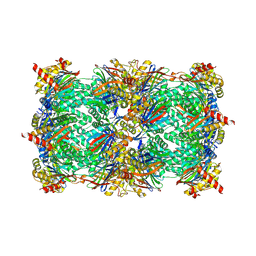 | | yCP beta5-A49V mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
