3DM8
 
 | | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris | | Descriptor: | DODECYL NONA ETHYLENE GLYCOL ETHER, uncharacterized protein RPA4348 | | Authors: | Cymborowski, M, Chruszcz, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris
To be Published
|
|
2GTA
 
 | | Crystal Structure of the putative pyrophosphatase YPJD from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR428. | | Descriptor: | Hypothetical protein ypjD, SODIUM ION | | Authors: | Vorobiev, S.M, Zhou, W, Seetharaman, J, Wang, D, Ma, L.C, Acton, T, Xio, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the putative pyrophosphatase YPJD from Bacillus subtilis.
To be Published
|
|
5UB9
 
 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni | | Descriptor: | ACETATE ION, ATP phosphoribosyltransferase, CHLORIDE ION, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
3DX2
 
 | | Golgi mannosidase II complex with MANNOSTATIN B | | Descriptor: | (1R,2R,3R,4S,5R)-4-amino-5-[(R)-methylsulfinyl]cyclopentane-1,2,3-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Kuntz, D.A, Zhong, W, Guo, J, Rose, D.R, Boons, G.-J. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
2GLL
 
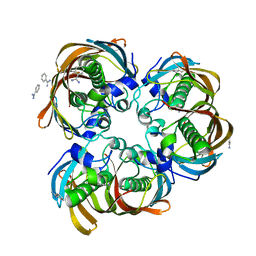 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GV7
 
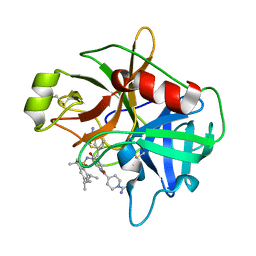 | | Structure of Matriptase in Complex with Inhibitor CJ-672 | | Descriptor: | (S)-4-(4-(3-(3-CARBAMIMIDOYLPHENYL)-2-(2,4,6-TRIISOPROPYLPHENYLSULFONAMIDO)PROPANOYL)PIPERAZINE-1-CARBONYL)PIPERIDINE-1-CARBOXIMIDAMIDE, Suppressor of tumorigenicity 14 | | Authors: | Bode, W. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Secondary Amides of Sulfonylated 3-Amidinophenylalanine. New Potent and Selective Inhibitors of Matriptase.
J.Med.Chem., 49, 2006
|
|
2C1X
 
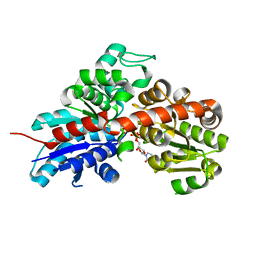 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
3BP7
 
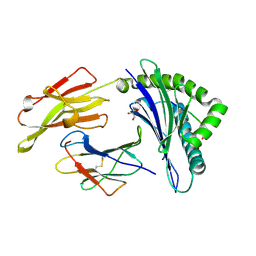 | | The high resolution crystal structure of HLA-B*2709 in complex with a Cathepsin A signal sequence peptide, pCatA | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
1ZQ7
 
 | | X-Ray Crystal Structure of Protein Q8PZK8 from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR9. | | Descriptor: | Hypothetical protein MM0484 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Yong, W, Xiao, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-18 | | Release date: | 2005-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-Ray structure of the hypothetical protein Q8PZK8 from Methanosarcina mazei at the resolution 2.1A. Northeast Structural Genomics Consortium target MaR9.
To be Published
|
|
1RS6
 
 | | Rat neuronal NOS heme domain with D-lysine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
5TSD
 
 | | Crystal structure of NADPH-dependent 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH and oxalate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALIC ACID, Probable hydroxyacid dehydrogenase protein | | Authors: | Matelska, D, Shabalin, I.G, Kutner, J, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of NADPH-dependent 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH and oxalate
to be published
|
|
1ZVI
 
 | | Rat Neuronal Nitric Oxide Synthase Oxygenase Domain | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric-oxide synthase, brain, ... | | Authors: | Matter, H, Kumar, H.S, Fedorov, R, Frey, A, Kotsonis, P, Hartmann, E, Frohlich, L.G, Reif, A, Pfleiderer, W, Scheurer, P, Ghosh, D.K, Schlichting, I, Schmidt, H.H. | | Deposit date: | 2005-06-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Isoform-Specific Inhibitors Targeting the Tetrahydrobiopterin Binding Site of Human Nitric Oxide Synthases.
J.Med.Chem., 48, 2005
|
|
5TT2
 
 | |
1ZVB
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
3BZZ
 
 | | Crystal structural of the mutated R313T EscU/SpaS C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.407 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
2H01
 
 | | PY00414- Plasmodium yoelii thioredoxin peroxidase I | | Descriptor: | 2-CYS PEROXIREDOXIN | | Authors: | Artz, J, Qiu, W, Min, J.R, Dong, A, Lew, J, Melone, M, Alam, Z, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-12 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of PY00414 - a Plasmodium yoelii thioredoxin peroxidase I
To be Published
|
|
4BWM
 
 | | KlenTaq mutant in complex with a RNA/DNA hybrid | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOCP)-3', 5'-R(*AP*AP*AP*GP*GP*GP*CP*GP*CP*CP*GP*UP*GP*GP*UP*C)-3', ... | | Authors: | Blatter, N, Bergen, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-07-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure and Function of an RNA-Reading Thermostable DNA Polymerase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2H2S
 
 | |
2H67
 
 | | NMR structure of human insulin mutant HIS-B5-ALA, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Liu, M, Hu, S.Q, Jia, W, Arvan, P, Weiss, M.A. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | A Conserved Histidine in Insulin Is Required for the Foldability of Human Proinsulin: Structure and function of an Alab5 analog.
J.Biol.Chem., 281, 2006
|
|
5GND
 
 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
5G1D
 
 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNDECAN-4, SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
1ZV8
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | ACETATE ION, CACODYLATE ION, E2 glycoprotein, ... | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
5GAI
 
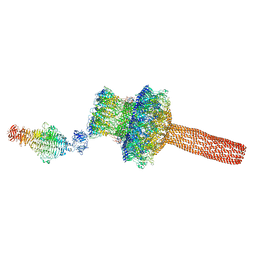 | | Probabilistic Structural Models of Mature P22 Bacteriophage Portal, Hub, and Tailspike proteins | | Descriptor: | Peptidoglycan hydrolase gp4, Portal protein, Tail fiber protein | | Authors: | Pintilie, G, Chen, D.H, Haase-Pettingell, C.A, King, J.A, Chiu, W. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage.
Biophys.J., 110, 2016
|
|
1ZR2
 
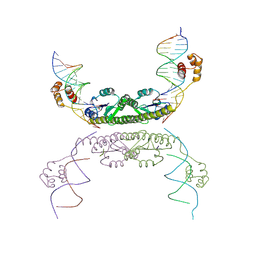 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently Linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
