7ARH
 
 | | LolCDE in complex with lipoprotein | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2RN5
 
 | | Humal Insulin Mutant B31Lys-B32Arg | | Descriptor: | Insulin | | Authors: | Bocian, W, Kozerski, L. | | Deposit date: | 2007-12-06 | | Release date: | 2008-10-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of biosynthetic engineered human insulin monomer B31(Lys)-B32(Arg) in water/acetonitrile solution. Comparison with the solution structure of native human insulin monomer
Biopolymers, 89, 2008
|
|
4XSR
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP-glucose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alr3699 protein, SULFATE ION, ... | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
3I12
 
 | | The crystal structure of the D-alanyl-alanine synthetase A from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase A | | Authors: | Zhang, R, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the D-alanyl-alanine synthetase A from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
4XTC
 
 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | Descriptor: | AlgM1, AlgM2, AlgQ2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
2STB
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2TSR
 
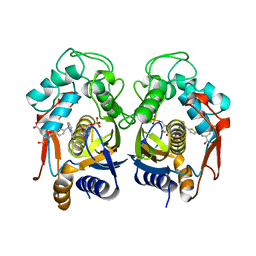 | | THYMIDYLATE SYNTHASE FROM RAT IN TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Sotelo-Mundo, R.R, Ciesla, J, Dzik, J.M, Rode, W, Maley, F, Maley, G, Hardy, L.W, Montfort, W.R. | | Deposit date: | 1998-06-19 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of rat thymidylate synthase inhibited by Tomudex, a potent anticancer drug.
Biochemistry, 38, 1999
|
|
4XZE
 
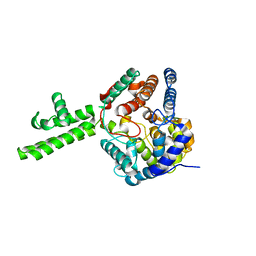 | | The crystal structure of Hazara virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
1X1I
 
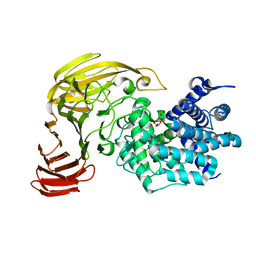 | | Crystal Structure of Xanthan Lyase (N194A) Complexed with a Product | | Descriptor: | (4AR,6R,7S,8R,8AS)-HEXAHYDRO-6,7,8-TRIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID, xanthan lyase | | Authors: | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-04-04 | | Release date: | 2005-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
7SP5
 
 | | Crystal Structure of a Eukaryotic Phosphate Transporter | | Descriptor: | PHOSPHATE ION, Phosphate transporter, nonyl beta-D-glucopyranoside | | Authors: | Stroud, R.M, Pedersen, B.P, Kumar, H, Waight, A.B, Risenmay, A.J, Roe-Zurz, Z, Chau, B.H, Schlessinger, A, Bonomi, M, Harries, W, Sali, A, Johri, A.K, Finer-Moore, J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a eukaryotic phosphate transporter.
Nature, 496, 2013
|
|
2R5X
 
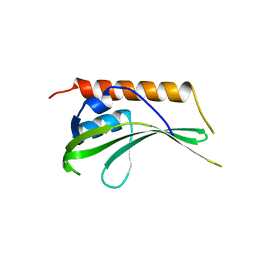 | | Crystal structure of uncharacterized conserved protein YugN from Geobacillus kaustophilus HTA426 | | Descriptor: | Uncharacterized conserved protein | | Authors: | Ramagopal, U.A, Patskovsky, Y, Shi, W, Toro, R, Meyer, A.J, Freeman, J, Wu, B, Koss, J, Groshong, C, Rodgers, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of uncharacterized protein YugN from Geobacillus kaustophilus HTA426.
To be Published
|
|
1DP6
 
 | | OXYGEN-BINDING COMPLEX OF FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
3HPB
 
 | |
7AEJ
 
 | |
2RA1
 
 | | Crystal structure of the N-terminal part of the bacterial S-layer protein SbsC | | Descriptor: | Surface layer protein | | Authors: | Pavkov, T, Egelseer, E.M, Tesarz, M, Sleytr, U.B, Keller, W. | | Deposit date: | 2007-09-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | The structure and binding behavior of the bacterial cell surface layer protein SbsC.
Structure, 16, 2008
|
|
2RJP
 
 | | Crystal structure of ADAMTS4 with inhibitor bound | | Descriptor: | ADAMTS-4, CALCIUM ION, N-({4'-[(4-isobutyrylphenoxy)methyl]biphenyl-4-yl}sulfonyl)-D-valine, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
7ARS
 
 | | The de novo designed hybrid alpha/beta-miniprotein (with Se-Methionine) | | Descriptor: | MAGNESIUM ION, alpha/beta-peptide, trifluoroacetic acid | | Authors: | Bejger, M, Fortuna, P, Drewniak-Switalska, M, Rypniewski, W, Berlicki, L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A computationally designed beta-amino acid-containing miniprotein.
Chem.Commun.(Camb.), 57, 2021
|
|
7ARR
 
 | | The de novo designed hybrid alpha/beta-miniprotein | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, alpha/beta-peptide, ... | | Authors: | Bejger, M, Fortuna, P, Drewniak-Switalska, M, Rypniewski, W, Berlicki, L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A computationally designed beta-amino acid-containing miniprotein.
Chem.Commun.(Camb.), 57, 2021
|
|
7SY4
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SY6
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417N mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SY7
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417T mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXT
 
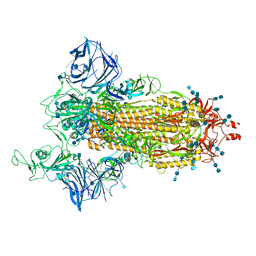 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXS
 
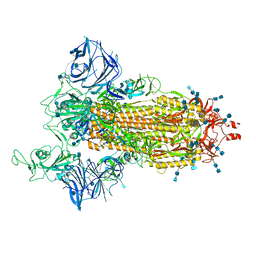 | | Cryo-EM structure of the SARS-CoV-2 D614G,L452R mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
4XSQ
 
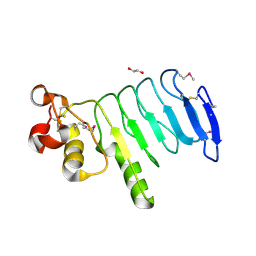 | | Structure of a variable lymphocyte receptor-like protein Bf66946 from Branchiostoma floridae | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, variable lymphocyte receptor-like protein Bf66946 | | Authors: | Cao, D.D, Cheng, W, Jiang, Y.L, Wang, W.J, Li, Q, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of a variable lymphocyte receptor-like protein from the amphioxus Branchiostoma floridae.
Sci Rep, 6, 2016
|
|
7SY5
 
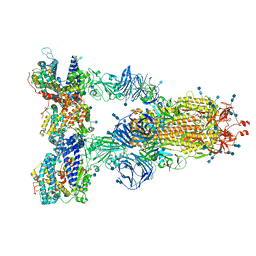 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417N mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
