5EMF
 
 | | Crystal structure of RNA r(GCUGCUGC) with antisense PNA p(GCAGCAGC) | | Descriptor: | CHLORIDE ION, RNA (5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3'), antisense PNA p(GCAGCAGC) | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
4ZSD
 
 | | Human Cyclophilin D Complexed with an Inhibitor at room temperature | | Descriptor: | 1-(4-aminobenzyl)-3-[(2S)-4-(methylsulfanyl)-1-{(2R)-2-[2-(methylsulfanyl)phenyl]pyrrolidin-1-yl}-1-oxobutan-2-yl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Delfosse, V, Allemand, F, Hoh, F, Sallaz-Damaz, Y, Pirocchi, M, Bourguet, W, Ferrer, J.-L, Labesse, G, Guichou, J.-F. | | Deposit date: | 2015-05-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2TSR
 
 | | THYMIDYLATE SYNTHASE FROM RAT IN TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Sotelo-Mundo, R.R, Ciesla, J, Dzik, J.M, Rode, W, Maley, F, Maley, G, Hardy, L.W, Montfort, W.R. | | Deposit date: | 1998-06-19 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of rat thymidylate synthase inhibited by Tomudex, a potent anticancer drug.
Biochemistry, 38, 1999
|
|
2R5X
 
 | | Crystal structure of uncharacterized conserved protein YugN from Geobacillus kaustophilus HTA426 | | Descriptor: | Uncharacterized conserved protein | | Authors: | Ramagopal, U.A, Patskovsky, Y, Shi, W, Toro, R, Meyer, A.J, Freeman, J, Wu, B, Koss, J, Groshong, C, Rodgers, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of uncharacterized protein YugN from Geobacillus kaustophilus HTA426.
To be Published
|
|
5A7Y
 
 | | Crystal structure of Sulfolobus acidocaldarius Trm10 in complex with S-adenosylhomocysteine | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
5AC6
 
 | | S.enterica HisA mutant D10G, dup13-15, Q24L, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | The Protein Structures, Functions and Dynamics of Adaptive Evolution
To be Published
|
|
1EXE
 
 | |
2RA1
 
 | | Crystal structure of the N-terminal part of the bacterial S-layer protein SbsC | | Descriptor: | Surface layer protein | | Authors: | Pavkov, T, Egelseer, E.M, Tesarz, M, Sleytr, U.B, Keller, W. | | Deposit date: | 2007-09-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | The structure and binding behavior of the bacterial cell surface layer protein SbsC.
Structure, 16, 2008
|
|
2RPP
 
 | | Solution structure of Tandem zinc finger domain 12 in Muscleblind-like protein 2 | | Descriptor: | Muscleblind-like protein 2, ZINC ION | | Authors: | Abe, C, Dang, W, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5EX0
 
 | | Crystal structure of human SMYD3 in complex with a MAP3K2 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, MAP3K2 peptide, ... | | Authors: | Fu, W, Liu, N, Qiao, Q, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
5AA8
 
 | | Structure of C1156Y,L1198F Mutant Human Anaplastic Lymphoma Kinase in Complex with PF-06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl- 15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y.-L, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
4HTC
 
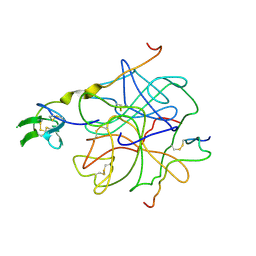 | | THE REFINED STRUCTURE OF THE HIRUDIN-THROMBIN COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Rydel, T.J, Bode, W, Huber, R. | | Deposit date: | 1993-06-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure of the hirudin-thrombin complex.
J.Mol.Biol., 221, 1991
|
|
2RGO
 
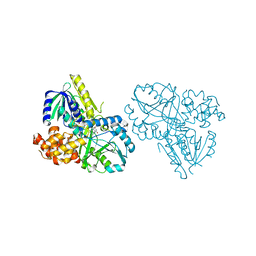 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
5F0W
 
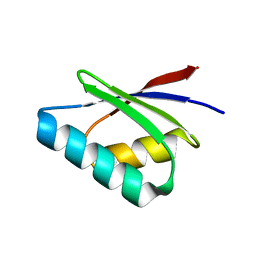 | |
5A4O
 
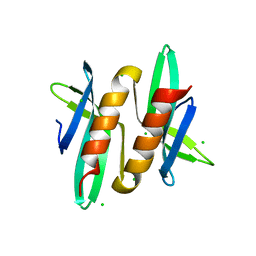 | |
7SP5
 
 | | Crystal Structure of a Eukaryotic Phosphate Transporter | | Descriptor: | PHOSPHATE ION, Phosphate transporter, nonyl beta-D-glucopyranoside | | Authors: | Stroud, R.M, Pedersen, B.P, Kumar, H, Waight, A.B, Risenmay, A.J, Roe-Zurz, Z, Chau, B.H, Schlessinger, A, Bonomi, M, Harries, W, Sali, A, Johri, A.K, Finer-Moore, J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a eukaryotic phosphate transporter.
Nature, 496, 2013
|
|
5A2P
 
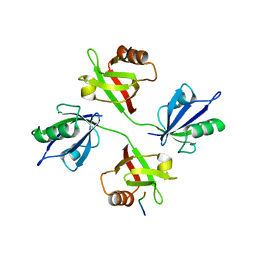 | |
5EWO
 
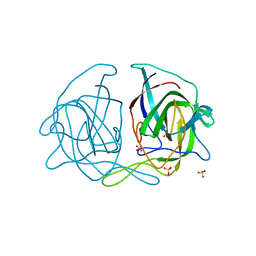 | | Crystal structure of the human astrovirus 1 capsid protein spike domain at 0.95-A resolution | | Descriptor: | SULFATE ION, Structural protein | | Authors: | Bogdanoff, W, York, R.L, Yousefi, P.A, Haile, S, Tripathi, S, DuBois, R.M. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural, Mechanistic, and Antigenic Characterization of the Human Astrovirus Capsid.
J.Virol., 90, 2015
|
|
2STA
 
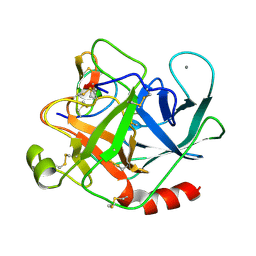 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA MAXIMA TRYPSIN INHIBITOR I) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-10 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5EX3
 
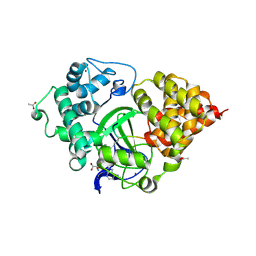 | | Crystal structure of human SMYD3 in complex with a VEGFR1 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Qiao, Q, Fu, W, Liu, N, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
2STB
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5EX4
 
 | | 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with tryptophan in all three allosteric binding sites | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Reichau, S, Jiao, W, Blackmore, N.J, Hutton, R.D, Parker, E.J. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with tryptophan in all three allosteric binding sites
to be published
|
|
7SY7
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417T mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SY4
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
