2OBM
 
 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, EscN | | Authors: | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1KHB
 
 | | PEPCK complex with nonhydrolyzable GTP analog, native data | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
2OCB
 
 | | Crystal structure of human RAB9B in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-9B, ... | | Authors: | Hong, B, Shen, L, Walker, J.R, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB9B in complex with a GTP analogue
To be Published
|
|
1KFI
 
 | | Crystal Structure of the Exocytosis-Sensitive Phosphoprotein, pp63/Parafusin (phosphoglucomutase) from Paramecium | | Descriptor: | SULFATE ION, ZINC ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-21 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
3R1E
 
 | | Crystal structure of GC(8BrG)GCGGC duplex | | Descriptor: | BROMIDE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.972 Å) | | Cite: | Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome.
Nucleic Acids Res., 39, 2011
|
|
3MYW
 
 | | The Bowman-Birk type inhibitor from mung bean in ternary complex with porcine trypsin | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Engh, R.A, Bode, W, Huber, R, Lin, G, Chi, C. | | Deposit date: | 2010-05-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 0.25-nm X-ray structure of the Bowman-Birk-type inhibitor from mung bean in ternary complex with porcine trypsin.
Eur.J.Biochem., 212, 1993
|
|
2OEN
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose-6-phosphate and fructose-1,6-bisphosphate | | Descriptor: | Catabolite control protein, Phosphocarrier protein HPr | | Authors: | Schumacher, M.A, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-12-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
1KM1
 
 | |
1KM5
 
 | |
3MXR
 
 | | SHV-1 beta-lactamase complex with compound 1 | | Descriptor: | ({[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-4-(4-hydroxyphenyl)butanoyl]amino}methyl)boronic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Novel Insights into the Mode of Inhibition of Class A SHV-1 {beta}-Lactamases Revealed by Boronic Acid Transition State Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3MX5
 
 | | Lassa fever virus nucleoprotein complexed with UTP | | Descriptor: | Nucleoprotein, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
2OCP
 
 | | Crystal Structure of Human Deoxyguanosine Kinase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxyguanosine kinase | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Substrate Specificities of Cellular Deoxyribonucleoside Kinases.
Nat.Struct.Biol., 8, 2001
|
|
2OK5
 
 | | Human Complement factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement factor B, ... | | Authors: | Milder, F.J, Gomes, L, Schouten, A, Janssen, B.J.C, Huizinga, E.G, Romijn, R.A, Hemrika, W, Roos, A, Daha, M.R, Gros, P. | | Deposit date: | 2007-01-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Factor B structure provides insights into activation of the central protease of the complement system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1IH8
 
 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2OGP
 
 | | Solution structure of the second PDZ domain of Par-3 | | Descriptor: | Partitioning-defective 3 homolog | | Authors: | Feng, W, Wu, H, Chen, J, Chan, L.-N, Zhang, M. | | Deposit date: | 2007-01-07 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | PDZ domains of par-3 as potential phosphoinositide signaling integrators
Mol.Cell, 28, 2007
|
|
1EJN
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN INHIBITOR COMPLEX | | Descriptor: | N-(1-ADAMANTYL)-N'-(4-GUANIDINOBENZYL)UREA, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Sperl, S, Jacob, U, Arroyo de Prada, N, Stuerzebecher, J, Wilhelm, O.G, Bode, W, Magdolen, V, Huber, R, Moroder, L. | | Deposit date: | 2000-04-22 | | Release date: | 2000-05-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | (4-aminomethyl)phenylguanidine derivatives as nonpeptidic highly selective inhibitors of human urokinase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3W25
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3Q5J
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
2NU2
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
3Q7J
 
 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
1JQG
 
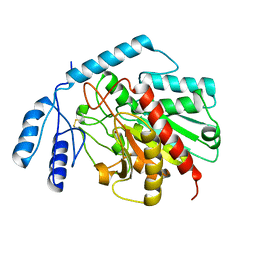 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
3EN0
 
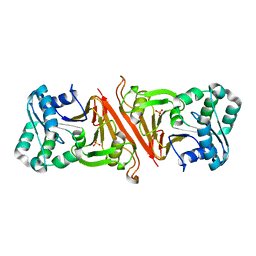 | | The Structure of Cyanophycinase | | Descriptor: | Cyanophycinase, SULFATE ION | | Authors: | Kimber, M.S, Law, A.M, Lai, S.W.S, Tavares, J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis of beta-peptide-specific cleavage by the serine protease cyanophycinase.
J.Mol.Biol., 392, 2009
|
|
1JY8
 
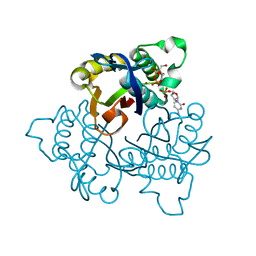 | | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2001-09-11 | | Release date: | 2002-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of 2C-methyl-d-erythritol-2,4-cyclodiphosphate synthase involved in mevalonate-independent biosynthesis of isoprenoids.
J.Mol.Biol., 316, 2002
|
|
3QTD
 
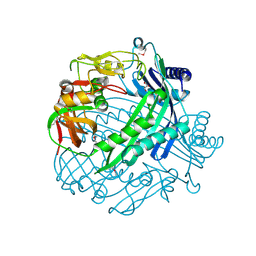 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
1KG1
 
 | | NMR structure of the NIP1 elicitor protein from Rhynchosporium secalis | | Descriptor: | Necrosis Inducing Protein 1 | | Authors: | Van't Slot, K.A, Van den Burg, H.A, Kloks, C.P, Hilbers, C.W, Knogge, W, Papavoine, C.H. | | Deposit date: | 2001-11-26 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Plant Disease Resistance-triggering Protein NIP1 from the Fungus Rhynchosporium secalis Shows a Novel beta-Sheet Fold.
J.Biol.Chem., 278, 2003
|
|
