6O0B
 
 | | Structural and Mechanistic Insights into CO2 Activation by Nitrogenase Iron Protein | | 分子名称: | IRON/SULFUR CLUSTER, Nitrogenase iron protein | | 著者 | Rettberg, L.A, Stiebritz, M.T, Kang, W, Lee, C.C, Ribbe, M.W, Hu, Y. | | 登録日 | 2019-02-15 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and Mechanistic Insights into CO2Activation by Nitrogenase Iron Protein.
Chemistry, 25, 2019
|
|
2NU2
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | 分子名称: | Ovomucoid, Streptogrisin B, Protease B | | 著者 | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | 登録日 | 2006-11-08 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
1XQV
 
 | | Crystal structure of inactive F1-mutant G37A | | 分子名称: | Proline iminopeptidase | | 著者 | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | 登録日 | 2004-10-13 | | 公開日 | 2005-07-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-17 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
1XR6
 
 | | Crystal Structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 1B | | 分子名称: | Genome polyprotein, POTASSIUM ION | | 著者 | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | 登録日 | 2004-10-13 | | 公開日 | 2004-10-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
1XRL
 
 | | Crystal structure of active site F1-mutant Y205F complex with inhibitor PCK | | 分子名称: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | 著者 | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | 登録日 | 2004-10-15 | | 公開日 | 2005-07-12 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRX
 
 | | Crystal structure of a DNA-binding protein | | 分子名称: | CALCIUM ION, SeqA protein | | 著者 | Guarne, A, Brendler, T, Zhao, Q, Ghirlando, R, Austin, S, Yang, W. | | 登録日 | 2004-10-16 | | 公開日 | 2005-05-10 | | 最終更新日 | 2013-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization.
Embo J., 24, 2005
|
|
1R5Q
 
 | |
1R78
 
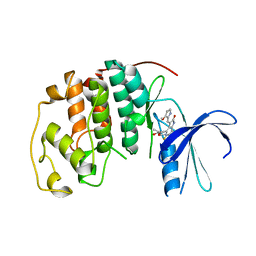 | | CDK2 complex with a 4-alkynyl oxindole inhibitor | | 分子名称: | 4-((3R,4S,5R)-4-AMINO-3,5-DIHYDROXY-HEX-1-YNYL)-5-FLUORO-3-[1-(3-METHOXY-1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Cell division protein kinase 2 | | 著者 | Luk, K.-C, Simcox, M.E, Schutt, A, Rowan, K, Thompson, T, Chen, Y, Kammlott, U, DePinto, W, Dunten, P, Dermatakis, A. | | 登録日 | 2003-10-20 | | 公開日 | 2004-01-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A new series of potent oxindole inhibitors of CDK2
Bioorg.Med.Chem.Lett., 14, 2004
|
|
3P1J
 
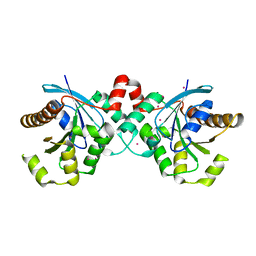 | | Crystal structure of human GTPase IMAP family member 2 in the nucleotide-free state | | 分子名称: | GTPase IMAP family member 2, UNKNOWN ATOM OR ION | | 著者 | Shen, L, Tempel, W, Tong, Y, Guan, X, Nedyalkova, L, Wernimont, A.K, Mackenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Andrews, D.W, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-09-30 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Crystal structure of human GTPase IMAP family member 2 in the nucleotide-free state
to be published
|
|
3P1W
 
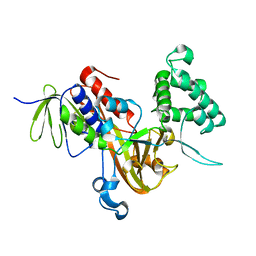 | | Crystal Structure of RAB GDI from Plasmodium Falciparum, PFL2060c | | 分子名称: | RabGDI protein | | 著者 | Wernimont, A.K, Neculai, A.M, Weadge, J, MacKenzie, F, Cossar, D, Tempel, W, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Langsley, G, Bosch, J, Hui, R, Pizzaro, J.C, Hutchinson, A, Structural Genomics Consortium (SGC) | | 登録日 | 2010-09-30 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of RAB GDI from Plasmodium Falciparum, PFL2060c
To be published
|
|
2O9Q
 
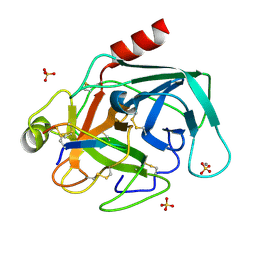 | | The crystal structure of Bovine Trypsin complexed with a small inhibition peptide ORB2K | | 分子名称: | CALCIUM ION, Cationic trypsin, ORB2K, ... | | 著者 | Li, J, Zhang, C, Xu, X, Wang, J, Gong, W, Lai, R. | | 登録日 | 2006-12-14 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | From protease inhibitor to antibiotics: single point mutation makes tremendous functional shift
To be Published
|
|
6NVX
 
 | |
1XRQ
 
 | | Crystal structure of active site F1-mutant E245Q soaked with peptide Phe-Leu | | 分子名称: | LEUCINE, Proline iminopeptidase | | 著者 | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | 登録日 | 2004-10-15 | | 公開日 | 2005-07-12 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
6NYF
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 1 (OA-1) | | 分子名称: | Vacuolating cytotoxin autotransporter | | 著者 | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | 登録日 | 2019-02-11 | | 公開日 | 2019-03-27 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3P8K
 
 | | Crystal Structure of a putative carbon-nitrogen family hydrolase from Staphylococcus aureus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gordon, R.D, Qiu, W, Battaile, K, Lam, K, Soloveychik, M, Benetteraj, D, Romanov, V, Pai, E.F, Chirgadze, N.Y. | | 登録日 | 2010-10-14 | | 公開日 | 2011-10-19 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of carbon-nitrogen family hydrolase from Staphylococcus aureus
To be Published
|
|
6O0V
 
 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-17 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6NRV
 
 | |
5S73
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | 分子名称: | Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
6NIM
 
 | | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine | | 分子名称: | Bromodomain factor 2 protein, Bromosporine, SULFATE ION, ... | | 著者 | Lin, Y.H, Dong, A, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R, Structural Genomics Consortium (SGC) | | 登録日 | 2018-12-29 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine
to be published
|
|
4P0Z
 
 | |
6NUE
 
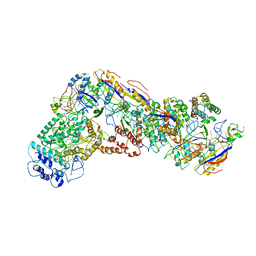 | | Small conformation of apo CRISPR_Csm complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | 著者 | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | 登録日 | 2019-01-31 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
5S74
 
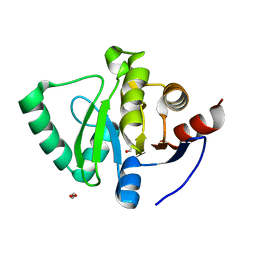 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
1R2W
 
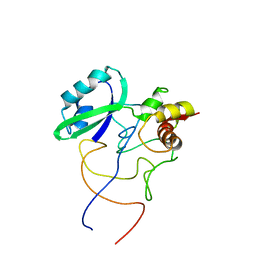 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of the 70S ribosome | | 分子名称: | 50S ribosomal protein L11, 58nts of 23S rRNA | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-30 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
3OQN
 
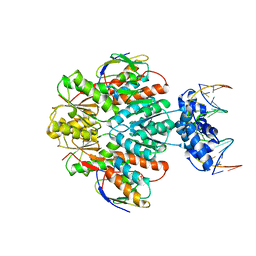 | | Structure of ccpa-hpr-ser46-p-gntr-down cre | | 分子名称: | 5'-D(*AP*TP*GP*GP*TP*AP*CP*CP*GP*CP*TP*TP*TP*CP*AP*A)-3', 5'-D(*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*TP*AP*CP*CP*AP*T)-3', Catabolite control protein A, ... | | 著者 | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | 登録日 | 2010-09-03 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
