3O47
 
 | | Crystal structure of ARFGAP1-ARF1 fusion protein | | Descriptor: | ADP-ribosylation factor GTPase-activating protein 1, ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Tong, Y, Nedyalkova, L, Tempel, W, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ARFGAP1-ARF1 fusion protein
to be published
|
|
5TRE
 
 | | Zinc and the Iron Donor Frataxin Regulate Oligomerization of the Scaffold Protein to Form New Fe-S Cluster Assembly Centers | | Descriptor: | Frataxin homolog, mitochondrial, Iron sulfur cluster assembly protein 1 | | Authors: | Ranatunga, W, Gakh, O, Galeano, B.K, Smith IV, D.Y, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-10-26 | | Release date: | 2017-06-07 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (15.6 Å) | | Cite: | Zinc and the iron donor frataxin regulate oligomerization of the scaffold protein to form new Fe-S cluster assembly centers.
Metallomics, 9, 2017
|
|
4MIW
 
 | | High-resolution structure of the N-terminal endonuclease domain of the Lassa virus L polymerase | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L | | Authors: | Wallat, G.D, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2013-09-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | High-resolution structure of the N-terminal endonuclease domain of the lassa virus L polymerase in complex with magnesium ions.
Plos One, 9, 2014
|
|
3NTV
 
 | | Crystal structure of a putative caffeoyl-CoA O-methyltransferase from Staphylococcus aureus | | Descriptor: | MW1564 protein, SULFATE ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-07-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a putative caffeoyl-CoA O-methyltransferase from Staphylococcus aureus
TO BE PUBLISHED
|
|
5YAZ
 
 | | Crystal structure of the ANKRD domain of KANK1 | | Descriptor: | ACETATE ION, KN motif and ankyrin repeat domains 1 | | Authors: | Wei, Z, Pan, W. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
5TV4
 
 | | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, L-glycero-alpha-D-manno-heptopyranose-(1-7)-L-glycero-alpha-D-manno-heptopyranose-(1-3)-L-glycero-alpha-D-manno-heptopyranose-(1-5)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-alpha-D-glucopyranose-(1-6)-2-amino-2-deoxy-alpha-D-glucopyranose, LAURIC ACID, ... | | Authors: | Mi, W, Walz, T, Liao, M. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of MsbA-mediated lipopolysaccharide transport.
Nature, 549, 2017
|
|
4MSV
 
 | | Crystal structure of FASL and DcR3 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 6, ... | | Authors: | Liu, W, Ramagopal, U.A, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
3O46
 
 | | Crystal structure of the PDZ domain of MPP7 | | Descriptor: | MAGUK p55 subfamily member 7, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Zhong, N, Guan, X, Landry, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the PDZ domain of MPP7
TO BE PUBLISHED
|
|
4V9I
 
 | | Crystal structure of thermus thermophilus 70S in complex with tRNAs and mRNA containing a pseudouridine in a stop codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S Ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
1M0O
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-methylpropanephosphonate | | Descriptor: | (1R)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]-1-METHYLPROPYLPHOSPHONIC ACID, 2,2-Dialkylglycine decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
3O1M
 
 | | Iron-Catalyzed Oxidation Intermediates Captured in A DNA Repair Dioxygenase | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, DNA (5'-D(*AP*AP*CP*GP*GP*TP*AP*TP*TP*AP*CP*CP*T)-3'), ... | | Authors: | Yi, C, Jia, G, Hou, G, Dai, Q, Zhang, W, Zheng, G, Jian, X, Yang, C.-G, Cui, Q, He, C. | | Deposit date: | 2010-07-21 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase.
Nature, 468, 2010
|
|
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
3O1V
 
 | | Iron-Catalyzed Oxidation Intermediates Captured in A DNA Repair Dioxygenase | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase AlkB, DNA (5'-D(*AP*AP*CP*GP*GP*TP*AP*TP*TP*AP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*TP*AP*AP*(MDJ)P*AP*CP*CP*GP*T)-3'), ... | | Authors: | Yi, C, Jia, G, Hou, G, Dai, Q, Zhang, W, Zheng, G, Jian, X, Yang, C.-G, Cui, Q, He, C. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase.
Nature, 468, 2010
|
|
4MV0
 
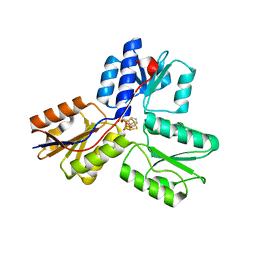 | | IspH in complex with pyridin-2-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-2-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
5UHF
 
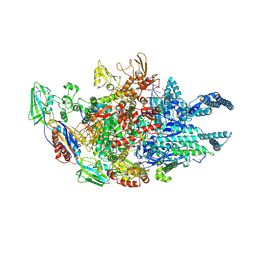 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with D-IX336 | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.345 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
3OBL
 
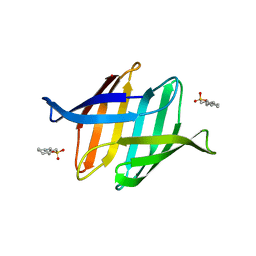 | |
4N4R
 
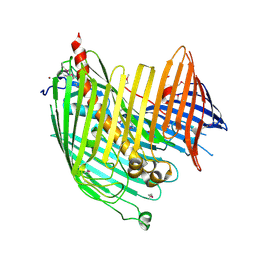 | | Structure basis of lipopolysaccharide biogenesis | | Descriptor: | CACODYLATE ION, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Dong, H, Xiang, Q, Wang, Z, Paterson, N.G, He, C, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-06-25 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for outer membrane lipopolysaccharide insertion.
Nature, 511, 2014
|
|
3O3N
 
 | | (R)-2-hydroxyisocaproyl-CoA dehydratase in complex with its substrate (R)-2-hydroxyisocaproyl-CoA | | Descriptor: | HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl ]oxy-hydroxy-phosphoryl]oxy-2-hydroxy-3,3-dimethyl-butanoyl]amino]propanoylamino]ethyl] (2R)-2-hydroxy-4-methyl-pentanethioate, ... | | Authors: | Knauer, S.H, Buckel, W, Dobbek, H. | | Deposit date: | 2010-07-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Reductive Radical Formation and Electron Recycling in (R)-2-Hydroxyisocaproyl-CoA Dehydratase.
J.Am.Chem.Soc., 133, 2011
|
|
5Y9J
 
 | | BAFF in complex with belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belibumab light chain, belimumab heavy chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
4N8C
 
 | | Three-dimensional structure of the extracellular domain of Matrix protein 2 of influenza A virus | | Descriptor: | Extracellular domain of influenza Matrix protein 2, Heavy chain of monoclonal antibody, Light chain of monoclonal antibody | | Authors: | Cho, K.J, Seok, J.H, Kim, S, Roose, K, Schepens, B, Fiers, W, Saelens, X, Kim, K.H. | | Deposit date: | 2013-10-17 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody
J.Virol., 89, 2015
|
|
5TX7
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Czub, M.P, Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Kutner, J, Cymborowski, M.T, Hennig, P.M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-11-15 | | Release date: | 2016-12-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris
to be published
|
|
1MKN
 
 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
3ZI8
 
 | | Structure of the R17A mutant of the Ralstonia soleanacerum lectin at 1.5 Angstrom in complex with L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, MAGNESIUM ION, PUTATIVE FUCOSE-BINDING LECTIN PROTEIN, ... | | Authors: | Arnaud, J, Audfray, A, Claudinon, J, Trondle, K, Trosvalet, M, Thomas, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reduction of Lectin Valency Drastically Changes Glycolipid Dynamics in Membranes, But not Surface Avidity.
Acs Chem.Biol., 8, 2013
|
|
5YEU
 
 | | Structural and mechanistic analyses reveal a unique Cas4-like protein in the mimivirus virophage resistance element system | | Descriptor: | MAGNESIUM ION, Uncharacterized protein R354 | | Authors: | Dou, C, Yu, M.J, Gu, Y.J, Cheng, W. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System.
Iscience, 3, 2018
|
|
5YF8
 
 | |
