5YEU
 
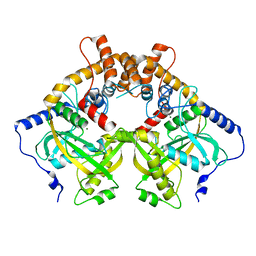 | | Structural and mechanistic analyses reveal a unique Cas4-like protein in the mimivirus virophage resistance element system | | Descriptor: | MAGNESIUM ION, Uncharacterized protein R354 | | Authors: | Dou, C, Yu, M.J, Gu, Y.J, Cheng, W. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System.
Iscience, 3, 2018
|
|
5YET
 
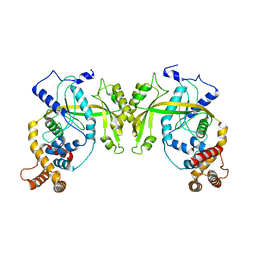 | | Structure of R354_WT | | Descriptor: | Uncharacterized protein R354 | | Authors: | Dou, C, Yu, M.J, Gu, Y.J, Cheng, W. | | Deposit date: | 2017-09-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System.
Iscience, 3, 2018
|
|
3SPL
 
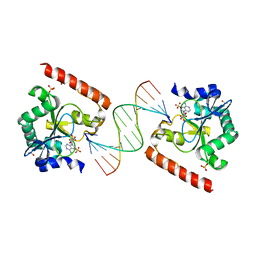 | | Crystal structure of aprataxin ortholog Hnt3 in complex with DNA and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin-like protein, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*GP*AP*G)-3'), ... | | Authors: | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | Deposit date: | 2011-07-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SPD
 
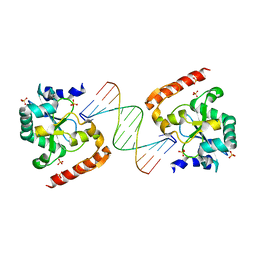 | | Crystal structure of aprataxin ortholog Hnt3 in complex with DNA | | Descriptor: | Aprataxin-like protein, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SP4
 
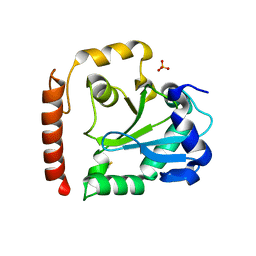 | | Crystal structure of aprataxin ortholog Hnt3 from Schizosaccharomyces pombe | | Descriptor: | Aprataxin-like protein, SULFATE ION, ZINC ION | | Authors: | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | Authors: | Liu, C, Song, D, Dou, C. | | Deposit date: | 2021-02-04 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
5ZW6
 
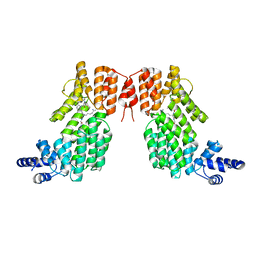 | | Structure of spAimR | | Descriptor: | AimR transcriptional regulator, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZVV
 
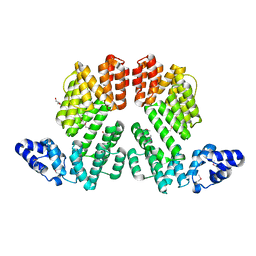 | | Structure of SeMet-phAimR | | Descriptor: | AimR transcriptional regulator, GLYCEROL | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW5
 
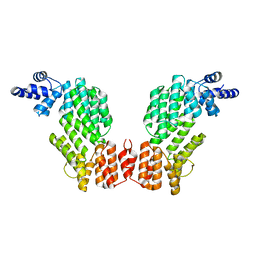 | | Structure of SeMet-spAimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-29 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZVW
 
 | | Structure of phAimR-Ligand | | Descriptor: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5XUO
 
 | |
