1XI6
 
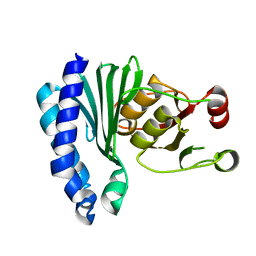 | | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001 | | Descriptor: | extragenic suppressor | | Authors: | Zhao, M, Chang, J.C, Zhou, W, Chen, L, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001
To be published
|
|
1TW0
 
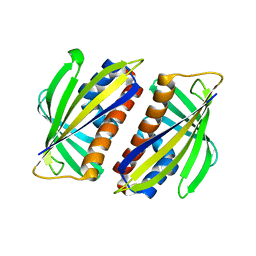 | |
2NWA
 
 | | X-ray Crystal Structure of Protein ytmB from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR466 | | Descriptor: | Hypothetical protein ytmB, SULFATE ION | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Wang, D, Cunningham, K, Ma, L.-C, Fang, Y, Xiao, R, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Hypothetical protein YtmB from Bacillus subtilis subsp. (subtilis str. 168), Northeast Structural Genomics target SR466
To be Published
|
|
1TX7
 
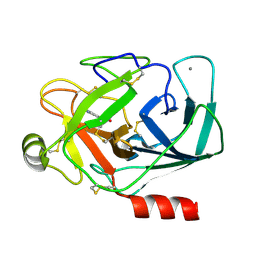 | | Bovine Trypsin complexed with p-amidinophenylmethylphosphinic acid (AMPA) | | Descriptor: | (4-CARBAMIMIDOYLPHENYL)-METHYL-PHOSPHINIC ACID, CALCIUM ION, Trypsinogen | | Authors: | Cui, J, Marankan, F, Fu, W, Crich, D, Mesecar, A, Johnson, M.E. | | Deposit date: | 2004-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An oxyanion-hole selective serine protease inhibitor in complex with trypsin.
Bioorg.Med.Chem., 10, 2002
|
|
4QVN
 
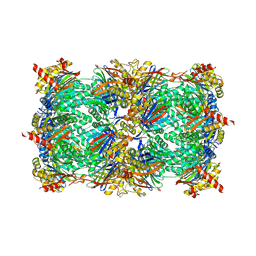 | | yCP beta5-M45V mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
2NSV
 
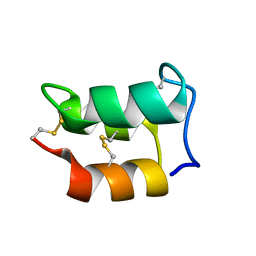 | |
6N7I
 
 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (gp4(5)-DNA) | | Descriptor: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
3MQ7
 
 | | Crystal Structure of Ectodomain Mutant of BST-2/Tetherin/CD317 | | Descriptor: | Bone marrow stromal antigen 2, CALCIUM ION | | Authors: | Xiong, Y, Yang, H, Wang, J, Meng, W. | | Deposit date: | 2010-04-27 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insight into the mechanisms of enveloped virus tethering by tetherin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4QWF
 
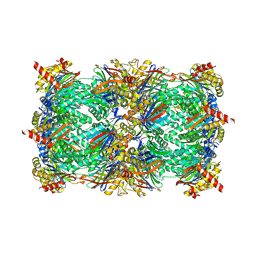 | | yCP beta5-M45I mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3MR3
 
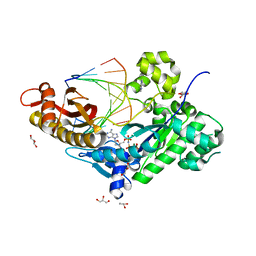 | | Human DNA polymerase eta - DNA ternary complex with the 3'T of a CPD in the active site (TT1) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*(TTD)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
6N9U
 
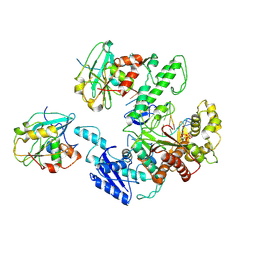 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) interacting with primase domains of two gp4 subunits bound to an RNA/DNA hybrid and dTTP (from LagS1) | | Descriptor: | DNA (44-MER), DNA primase/helicase, DNA-directed DNA polymerase, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
1XB4
 
 | |
6MH8
 
 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
4QYN
 
 | | The Crystal Structures of holo-wt human Cellular Retinol Binding protein II (hCRBPII) bound to Retinol | | Descriptor: | ACETATE ION, RETINOL, Retinol-binding protein 2 | | Authors: | Nossoni, Z, Assar, Z, Yapici, I, Nosrati, M, Wang, W, Berbasova, T, Vasileiou, C, Borhan, B, Geiger, H. | | Deposit date: | 2014-07-24 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structures of holo wild-type human cellular retinol-binding protein II (hCRBPII) bound to retinol and retinal.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1UCT
 
 | | Crystal structure of the extracellular fragment of Fc alpha Receptor I (CD89) | | Descriptor: | Immunoglobulin alpha Fc receptor | | Authors: | Ding, Y, Xu, G, Yang, M, Zhang, W, Rao, Z. | | Deposit date: | 2003-04-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Ectodomain of Human Fc{alpha}RI.
J.Biol.Chem., 278, 2003
|
|
2OBL
 
 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | Descriptor: | ACETATE ION, CALCIUM ION, EscN, ... | | Authors: | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6MKI
 
 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the ceftaroline-bound form | | Descriptor: | Ceftaroline, bound form, GLYCEROL, ... | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4CUO
 
 | | Banyan peroxidase with glycosylation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BANYAN PEROXIDASE, CALCIUM ION, ... | | Authors: | Palm, G.J, Sharma, A, Hinrichs, W. | | Deposit date: | 2014-03-20 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Post-Translational Modification and Extended Glycosylation Pattern of a Plant Latex Peroxidase of Native Source Characterized by X-Ray Crystallography.
FEBS J., 281, 2014
|
|
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | Descriptor: | penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4RBL
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with Mitoxantrone derivatives | | Descriptor: | 1,4,5,8-tetrahydroxyanthracene-9,10-dione, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Huang, N. | | Deposit date: | 2014-09-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Ser/Thr kinase Pim1 in complex with Mitoxantrone derivatives
To be Published, 2014
|
|
2OIE
 
 | | Crystal structure of RS21-C6 core segment RSCUT | | Descriptor: | RS21-C6, SULFATE ION | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2OKX
 
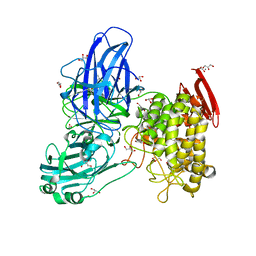 | | Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A | | Descriptor: | CALCIUM ION, GLYCEROL, Rhamnosidase B | | Authors: | Cui, Z, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase Family 78 alpha-L-Rhamnosidase from Bacillus sp. GL1
J.Mol.Biol., 374, 2007
|
|
3M6L
 
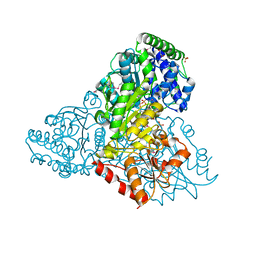 | | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion
To be Published
|
|
4RE8
 
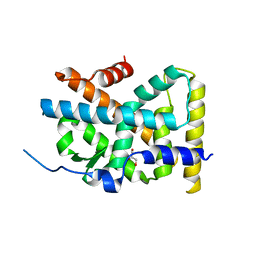 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
1U1E
 
 | | Structure of e. coli uridine phosphorylase complexed to 5(phenylseleno)acyclouridine (PSAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLSELANYL)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
