2GH4
 
 | | YteR/D143N/dGalA-Rha | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-03-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate
Biochem.Biophys.Res.Commun., 347, 2006
|
|
5EX4
 
 | | 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with tryptophan in all three allosteric binding sites | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Reichau, S, Jiao, W, Blackmore, N.J, Hutton, R.D, Parker, E.J. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with tryptophan in all three allosteric binding sites
to be published
|
|
3C7X
 
 | | Hemopexin-like domain of matrix metalloproteinase 14 | | Descriptor: | CHLORIDE ION, Matrix metalloproteinase-14, SODIUM ION | | Authors: | Tochowicz, A, Itoh, Y, Maskos, K, Bode, W, Goettig, P. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions
J.Biol.Chem., 286, 2011
|
|
4NX9
 
 | | Crystal structure of Pseudomonas aeruginosa flagellin FliC | | Descriptor: | Flagellin | | Authors: | Song, W.S, Yoon, S.I. | | Deposit date: | 2013-12-09 | | Release date: | 2014-01-29 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament
Biochem.Biophys.Res.Commun., 444, 2014
|
|
3CDK
 
 | | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis | | Descriptor: | Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit A, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B | | Authors: | Kim, Y, Zhou, M, Stols, L, Eschenfeldt, W, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis.
To be Published
|
|
2G8K
 
 | |
2C6C
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with GPI-18431 (S)-2-(4-iodobenzylphosphonomethyl)-pentanedioic acid | | Descriptor: | (2S)-2-{[HYDROXY(4-IODOBENZYL)PHOSPHORYL]METHYL}PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Barinka, C, Li, W, Tsukamoto, T, Majer, P, Slusher, B.S, Konvalinka, J, Hilgenfeld, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Glutamate Carboxypeptidase II, a Drug Target in Neuronal Damage and Prostate Cancer.
Embo J., 25, 2006
|
|
2GF4
 
 | | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14 | | Descriptor: | ACETATE ION, CALCIUM ION, Protein Vng1086c | | Authors: | Benach, J, Zhou, W, Jayaraman, S, Forouhar, F.F, Janjua, H, Xiao, R, Ma, L.-C, Cunningham, K, Wang, D, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14
To be Published
|
|
2BU5
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
5EWN
 
 | | Crystal structure of the human astrovirus 1 capsid protein core domain at 2.6 A resolution | | Descriptor: | CHLORIDE ION, Structural protein | | Authors: | York, R.L, Yousefi, P.A, Bogdanoff, W, Haile, S, Tripathi, S, DuBois, R.M. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-23 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural, Mechanistic, and Antigenic Characterization of the Human Astrovirus Capsid.
J.Virol., 90, 2015
|
|
2C1Z
 
 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
4NJ4
 
 | | Crystal Structure of Human ALKBH5 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation.
Nucleic Acids Res., 42, 2014
|
|
2G6H
 
 | | Structure of rat nNOS heme domain (BH4 bound) in the reduced form | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Igarashi, J, Jamal, J, Yang, W, Poulos, T.L. | | Deposit date: | 2006-02-24 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of constitutive nitric oxide synthases with diatomic ligands bound.
J.Biol.Inorg.Chem., 11, 2006
|
|
2BU6
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | (N-{4-[(ETHYLANILINO)SULFONYL]-2-METHYLPHENYL}-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
3CF4
 
 | | Structure of the CODH component of the M. barkeri ACDS complex | | Descriptor: | ACETIC ACID, Acetyl-CoA decarboxylase/synthase alpha subunit, Acetyl-CoA decarboxylase/synthase epsilon subunit, ... | | Authors: | Gong, W, Hao, B, Wei, Z, Ferguson Jr, D.J, Tallant, T, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2008-03-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the alpha2 epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarboxylase/synthase complex
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7V4Y
 
 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
2G6J
 
 | | Structure of rat nNOS (L337N) heme domain (4-aminobiopterin bound) complexed with NO | | Descriptor: | (1S,2S)-1-(2,4-DIAMINOPTERIDIN-6-YL)PROPANE-1,2-DIOL, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Igarashi, J, Jamal, J, Yang, W, Poulos, T.L. | | Deposit date: | 2006-02-24 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of constitutive nitric oxide synthases with diatomic ligands bound.
J.Biol.Inorg.Chem., 11, 2006
|
|
2G8H
 
 | |
2G8Y
 
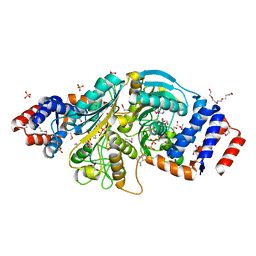 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
3CRR
 
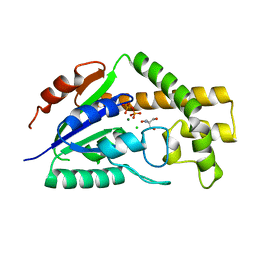 | | Structure of tRNA Dimethylallyltransferase: RNA Modification through a Channel | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Huang, R.H, Xie, W, Zhou, C. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of tRNA Dimethylallyltransferase: RNA Modification
through a Channel
J.Mol.Biol., 367, 2007
|
|
2C1C
 
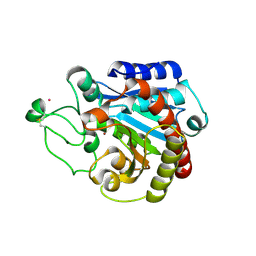 | | Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors | | Descriptor: | CARBOXYPEPTIDASE B, YTTRIUM ION, ZINC ION | | Authors: | Bayes, A, Comellas-Bigler, M, Rodriguez de la Vega, M, Maskos, K, Bode, W, Aviles, F.X, Jongsma, M.A, Beekwilder, J, Vendrell, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Resistance of an Insect Carboxypeptidase to Plant Protease Inhibitors.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5TF2
 
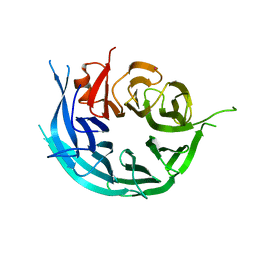 | | CRYSTAL STRUCTURE OF THE WD40 DOMAIN OF THE HUMAN PROLACTIN REGULATORY ELEMENT-BINDING PROTEIN | | Descriptor: | Prolactin regulatory element-binding protein, UNKNOWN ATOM OR ION | | Authors: | Walker, J.R, Zhang, Q, Dong, A, Wernimont, A, Li, Y, He, H, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Chen, Z, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-23 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF THE WD40 DOMAIN OF THE HUMAN PROLACTIN REGULATORY ELEMENT-BINDING PROTEIN (CASP target)
To be published
|
|
4CVH
 
 | | Crystal structure of human isoprenoid synthase domain-containing protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ISOPRENOID SYNTHASE DOMAIN-CONTAINING PROTEIN, ... | | Authors: | Kopec, J, Froese, D.S, Krojer, T, Newman, J, Kiyani, W, Goubin, S, Strain-Damerell, C, Vollmar, M, von Delft, F, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Lefeber, D.J, Yue, W.W. | | Deposit date: | 2014-03-27 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Human Ispd is a Cytidyltransferase Required for Dystroglycan O-Mannosylation.
Chem.Biol., 22, 2015
|
|
2BU7
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
5F0U
 
 | | Crystal structure of Gold binding protein | | Descriptor: | Putative copper chaperone, SILVER ION | | Authors: | Wei, W, Wang, F, Zhao, J. | | Deposit date: | 2015-11-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of tetrasilver bound to GolB at 1.70 Angstroms resolution
To Be Published
|
|
