3ET1
 
 | |
7V4Y
 
 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
1ORW
 
 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | Descriptor: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
3F5V
 
 | | C2 Crystal form of mite allergen DER P 1 | | Descriptor: | CALCIUM ION, Der p 1 allergen, HEXAETHYLENE GLYCOL | | Authors: | Stura, E.A, Minor, W, Chruszcz, M, Saint Remy, J.M. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of mite allergens Der f 1 and Der p 1 reveal differences in surface-exposed residues that may influence antibody binding.
J.Mol.Biol., 386, 2009
|
|
3I4P
 
 | | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cymborowski, M, Chruszcz, M, Luo, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens
To be Published
|
|
3VGZ
 
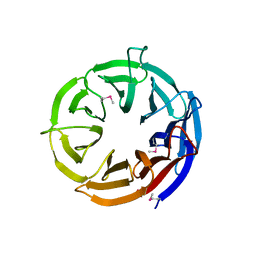 | | Crystal structure of E. coli YncE | | Descriptor: | Uncharacterized protein YncE | | Authors: | Kagawa, W, Sagawa, T, Niki, H, Kurumizaka, H. | | Deposit date: | 2011-08-23 | | Release date: | 2011-11-16 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the DNA-binding activity of the bacterial beta-propeller protein YncE
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3HS9
 
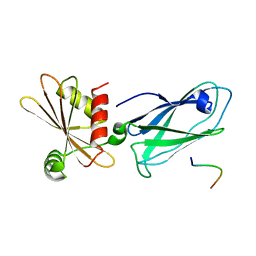 | | Intersectin 1-peptide-AP2 beta ear complex | | Descriptor: | AP-2 complex subunit beta-1, peptide from Intersectin-1, residues 841-851 | | Authors: | Vahedi-Faridi, A, Pechstein, A, Schaefer, J.G, Saenger, W, Haucke, V. | | Deposit date: | 2009-06-10 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3EZR
 
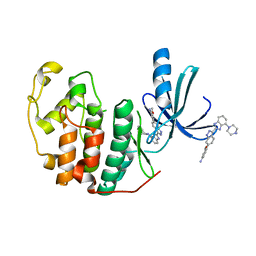 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1OPX
 
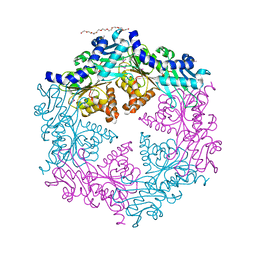 | | Crystal structure of the traffic ATPase (HP0525) of the Helicobacter pylori type IV secretion system bound by sulfate | | Descriptor: | NONAETHYLENE GLYCOL, SULFATE ION, virB11 homolog | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
3VLK
 
 | |
3F2R
 
 | | Crystal structure of human choline kinase alpha in complex with hemicholinium-3 | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), Choline kinase alpha, UNKNOWN ATOM OR ION | | Authors: | Hong, B, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of human choline kinase alpha in complex with hemicholinium-3
To be Published
|
|
1OT3
 
 | | Crystal structure of Drosophila deoxyribonucleotide kinase complexed with the substrate deoxythymidine | | Descriptor: | Deoxyribonucleoside Kinase, SULFATE ION, THYMIDINE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-21 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway: Studies of the Drosophila Deoxyribonucleoside Kinase.
Biochemistry, 42, 2003
|
|
3VPV
 
 | |
3HXB
 
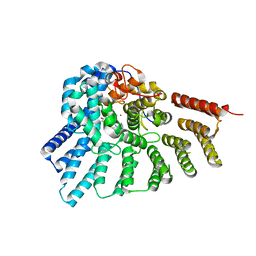 | | Engineered RabGGTase in complex with a peptidomimetic inhibitor (compound 6) | | Descriptor: | Geranylgeranyl transferase type-2 subunit alpha, Geranylgeranyl transferase type-2 subunit beta, N-[(benzyloxy)carbonyl]-D-tyrosyl-L-phenylalanyl-L-tyrosine, ... | | Authors: | Guo, Z, Alexandrov, K, Waldmann, H, Goody, R.S, Blankenfeldt, W. | | Deposit date: | 2009-06-20 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, synthesis, and characterization of Peptide-based rab geranylgeranyl transferase inhibitors
J.Med.Chem., 52, 2009
|
|
2JUV
 
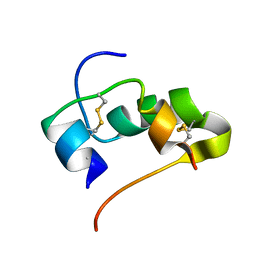 | | AbaA3-DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
3F7T
 
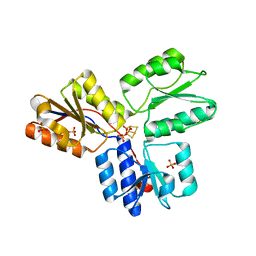 | | Structure of active IspH shows a novel fold with a [3Fe-4S] cluster in the catalytic centre | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PHOSPHATE ION, ... | | Authors: | Graewert, T, Eppinger, J, Rohdich, F, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2008-11-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3F1J
 
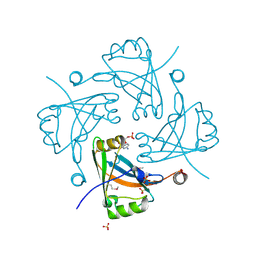 | | Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals RNA binding properties | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Matrix protein, SULFATE ION | | Authors: | Neumann, P, Lieber, D, Meyer, S, Dautel, P, Kerth, A, Kraus, I, Garten, W, Stubbs, M.T. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2JPC
 
 | |
3FK2
 
 | | Crystal structure of the RhoGAP domain of human glucocorticoid receptor DNA-binding factor 1 | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-15 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RhoGAP domain of human glucocorticoid receptor DNA-binding factor 1
To be Published
|
|
3HTQ
 
 | | the hemagglutinin structure of an avian H1N1 influenza A virus in complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
7YOA
 
 | |
3F5K
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
2K20
 
 | |
3HXI
 
 | | Crystal structure of Schistosome eIF4E complexed with m7GpppG and 4E-BP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Eukaryotic Translation Initiation 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Liu, W, Zhao, R, Jones, D.N.M, Davis, R.E. | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
J.Biol.Chem., 284, 2009
|
|
