8R4B
 
 | |
5DWS
 
 | | Crystal Structure of ITCH WW3 domain in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, UNKNOWN ATOM OR ION, txnip | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of ITCH WW3 domain in complex with TXNIP peptide
to be published
|
|
1Z3T
 
 | |
5DZD
 
 | | Crystal Structure of WW4 domain of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of WW4 domain of ITCH in complex with TXNIP peptide
To be Published
|
|
6WBK
 
 | |
5E1B
 
 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5DOX
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
2ORB
 
 | | The structure of the anti-c-myc antibody 9E10 Fab fragment | | Descriptor: | Monoclonal anti-c-myc antibody 9E10, SULFATE ION | | Authors: | Krauss, N, Scheerer, P, Hoehne, W. | | Deposit date: | 2007-02-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the anti-c-myc antibody 9E10 Fab fragment/epitope peptide complex reveals a novel binding mode dominated by the heavy chain hypervariable loops.
Proteins, 73, 2008
|
|
6VJV
 
 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
7BV5
 
 | | Crystal structure of the yeast heterodimeric ADAT2/3 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase subunit TAD2, tRNA-specific adenosine deaminase subunit TAD3 | | Authors: | Xie, W, Liu, X, Chen, R, Sun, Y, Chen, R, Zhou, J, Tian, Q. | | Deposit date: | 2020-04-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the yeast heterodimeric ADAT2/3 deaminase.
Bmc Biol., 18, 2020
|
|
6VIX
 
 | | BRD4_Bromodomain2 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6W9U
 
 | | Structure of human MAIT A-F7 TCR in complex with patient MR1-R9H-Ac-6-FP | | Descriptor: | 1-[[6-(1-$l^{1}-oxidanylethyl)-4-$l^{3}-oxidanylidene-2,3,6,8~{a}-tetrahydropteridin-2-yl]-$l^{2}-azanyl]ethanone, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1 .
Sci Immunol, 5, 2020
|
|
6TU9
 
 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
5EME
 
 | | Complex of RNA r(GCAGCAGC) with antisense PNA p(CTGCTGC) | | Descriptor: | Antisense PNA strand, CHLORIDE ION, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
5EPK
 
 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5END
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG)(Q152A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, GLYCEROL | | Authors: | Hou, J, Cooper, D.R, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
5EPE
 
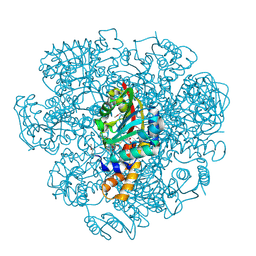 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
5EPJ
 
 | | Crystal Structure of chromodomain of CBX7 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 7, UNKNOWN ATOM OR ION, peptide-like inhibitor UNC3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-16 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of chromodomain of CBX7 in complex with inhibitor UNC3866
to be published
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
6TV2
 
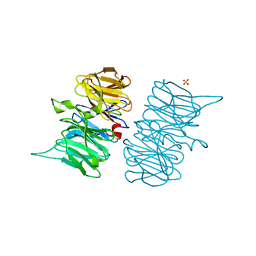 | | Heme d1 biosynthesis associated Protein NirF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | Authors: | Kluenemann, T, Layer, G, Blankenfeldt, W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
6TYD
 
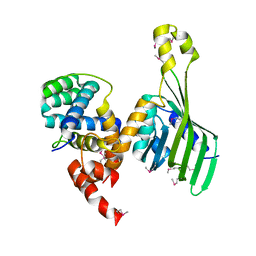 | | Structure of human LDB1 in complex with SSBP2 | | Descriptor: | LIM domain-binding protein 1, Single-stranded DNA-binding protein 2 | | Authors: | Wang, H, Wang, Z, Xu, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of human LDB1 in complex with SSBP2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5H1K
 
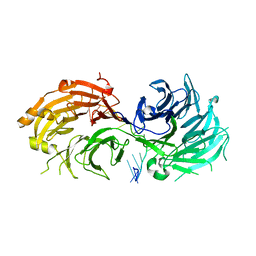 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 13-nt U4 snRNA fragment | | Descriptor: | Gem-associated protein 5, U4 snRNA (5'-R(*GP*CP*AP*AP*UP*UP*UP*UP*UP*GP*AP*CP*A)-3') | | Authors: | Wang, Y, Jin, W, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1M
 
 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with M7G | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H5D
 
 | |
8JTN
 
 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 2-propyl-2-azoniatricyclo[7.3.0.0^{3,7}]dodeca-1(9),2,7-trien-8-amine, Tudor domain-containing protein 3 | | Authors: | Chen, M, Wang, Z, Li, W, Shang, X, Liu, Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Tudor domain of TDRD3 in complex with a small molecule antagonist.
Biochim Biophys Acta Gene Regul Mech, 1866, 2023
|
|
