1EE1
 
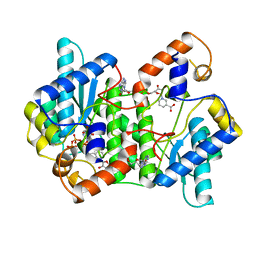 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE ATP, TWO MOLECULES DEAMIDO-NAD+ AND ONE MG2+ ION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, Delucas, L. | | Deposit date: | 2000-01-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ECA
 
 | |
3QT2
 
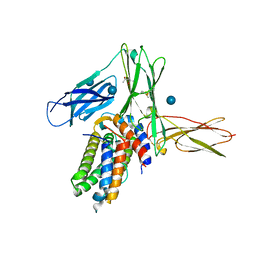 | | Structure of a cytokine ligand-receptor complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Interleukin-5, Interleukin-5 receptor subunit alpha, ... | | Authors: | Mueller, T.D, Patino, E, Kotzsch, A, Saremba, S, Nickel, J, Schmitz, W, Sebald, W. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure Analysis of the IL-5 Ligand-Receptor Complex Reveals a Wrench-like Architecture for IL-5Ralpha.
Structure, 19, 2011
|
|
1LWH
 
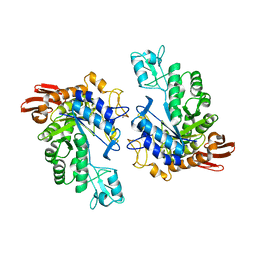 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE | | Descriptor: | 4-alpha-glucanotransferase, CALCIUM ION | | Authors: | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | Deposit date: | 2002-05-31 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
3N2T
 
 | | Structure of the glycerol dehydrogenase AKR11B4 from Gluconobacter oxydans | | Descriptor: | Putative oxidoreductase | | Authors: | Richter, N, Breicha, K, Hummel, W, Niefind, K. | | Deposit date: | 2010-05-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Three-Dimensional Structure of AKR11B4, a Glycerol Dehydrogenase from Gluconobacter oxydans, Reveals a Tryptophan Residue as an Accelerator of Reaction Turnover.
J.Mol.Biol., 404, 2010
|
|
2KRL
 
 | | The ensemble of the solution global structures of the 102-nt ribosome binding structure element of the turnip crinkle virus 3' UTR RNA | | Descriptor: | RNA (102-MER) | | Authors: | Zuo, X, Wang, J, Yu, P, Eyler, D, Xu, H, Starich, M, Tiede, D, Simon, A, Kasprzak, W, Schwieters, C, Shapiro, B. | | Deposit date: | 2009-12-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1REU
 
 | | Structure of the bone morphogenetic protein 2 mutant L51P | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, bone morphogenetic protein 2 | | Authors: | Keller, S, Nickel, J, Zhang, J.-L, Sebald, W, Mueller, T.D. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular recognition of BMP-2 and BMP receptor IA.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3NCR
 
 | | GlnK2 from Archaeoglubus fulgidus, ADP complex | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Helfmann, S, Lue, W, Litz, C, Andrade, S.L.A. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Cooperative binding of MgATP and MgADP in the trimeric P(II) protein GlnK2 from Archaeoglobus fulgidus.
J.Mol.Biol., 402, 2010
|
|
1MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
3PHY
 
 | | PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED), SOLUTION STRUCTURE, NMR, 26 STRUCTURES | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Dux, P, Rubinstenn, G, Vuister, G.W, Boelens, R, Mulder, F.A.A, Hard, K, Hoff, W.D, Kroon, A, Crielaard, W, Hellingwerf, K.J, Kaptein, R. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the photoactive yellow protein.
Biochemistry, 37, 1998
|
|
3PZW
 
 | | Soybean lipoxygenase-1 - re-refinement | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | Authors: | Chruszcz, M, Minor, W. | | Deposit date: | 2010-12-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Determination of protein structures - a series of fortunate events.
Biophys.J., 95, 2008
|
|
4J4X
 
 | | Crystal structure of GraVN | | Descriptor: | NP protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
3UGF
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
4BWJ
 
 | | KlenTaq mutant in complex with DNA and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOCP)-3', ... | | Authors: | Blatter, N, Bergen, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-07-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and Function of an RNA-Reading Thermostable DNA Polymerase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1N56
 
 | | Y-family DNA polymerase Dpo4 in complex with DNA containing abasic lesion | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*AP*A)-3', 5'-D(*TP*CP*AP*TP*(3DR)P*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2002-11-04 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts.
Mol.Cell, 13, 2004
|
|
4ISW
 
 | | Crystal Structure of Phosphorylated C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-17 | | Release date: | 2013-12-11 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Bioorg.Chem., 52C, 2013
|
|
4IW7
 
 | | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis. | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Newcomb, W, Niedzialkowska, E, Porebski, P.J, Grimshaw, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis.
To be Published
|
|
4IXU
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 11d: {(5R)-5-amino-5-carboxy-5-[(3-endo)-8-(3,4-dichlorobenzyl)-8-azabicyclo[3.2.1]oct-3-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3UV7
 
 | | Ec_IspH in complex with buta-2,3-dienyl diphosphate (1300) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, buta-2,3-dien-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UV3
 
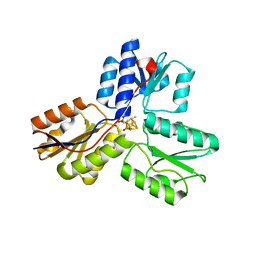 | | Ec_IspH in complex with but-2-ynyl diphosphate (1086) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, but-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
4BWE
 
 | | Crystal structure of C-terminally truncated glypican-1 after controlled dehydration to 86 percent relative humidity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glypican-1 | | Authors: | Awad, W, Svensson Birkedal, G, Thunnissen, M.M.G.M, Mani, K, Logan, D.T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Improvements in the order, isotropy and electron density of glypican-1 crystals by controlled dehydration.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
3NCQ
 
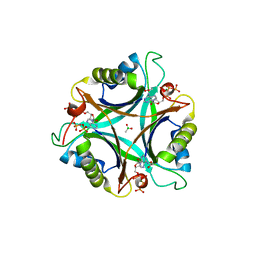 | | GlnK2 from Archaeoglobus fulgidus, ATP complex | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Helfmann, S, Lue, W, Litz, C, Andrade, S.L.A. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Cooperative binding of MgATP and MgADP in the trimeric P(II) protein GlnK2 from Archaeoglobus fulgidus.
J.Mol.Biol., 402, 2010
|
|
3UTD
 
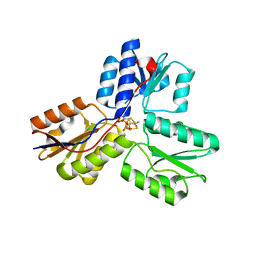 | | Ec_IspH in complex with 4-oxopentyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-oxopentyl trihydrogen diphosphate, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
1NDA
 
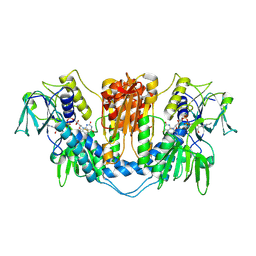 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
4L7G
 
 | | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxy-benzylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against Beta-Secretase-1 (BACE1) | | Descriptor: | (3aS,7aR)-7a-[3-(pyrimidin-5-yl)phenyl]-3a,6,7,7a-tetrahydro-4H-pyrano[4,3-d][1,3]oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE | | Authors: | Huestis, M.P, Liu, W, Volgraf, M, Purkey, H, Wang, W, Yu, C, Wu, P, Smith, D, Vigers, G, Dutcher, D, Geck Do, M.K, Hunt, K.W, Siu, M. | | Deposit date: | 2013-06-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxycycloalkylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against β-Secretase-1 (BACE-1)
Tetrahedron Lett., 2013
|
|
