3CIF
 
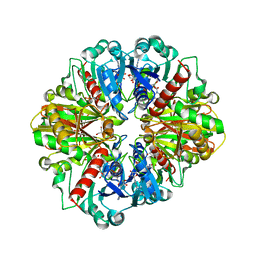 | | Crystal Structure of C153S mutant glyceraldehyde 3-phosphate dehydrogenase from Cryptosporidium parvum | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Cook, W.J, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An unexpected phosphate binding site in Glyceraldehyde 3-Phosphate Dehydrogenase: Crystal structures of apo, holo and ternary complex of Cryptosporidium parvum enzyme
BMC STRUCT.BIOL., 9, 2009
|
|
3CQF
 
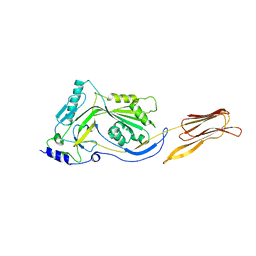 | | Crystal structure of anthrolysin O (ALO) | | Descriptor: | Thiol-activated cytolysin | | Authors: | Bourdeau, R.W, Malito, E, Tang, W.J. | | Deposit date: | 2008-04-02 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cellular Functions and X-ray Structure of Anthrolysin O, a Cholesterol-dependent Cytolysin Secreted by Bacillus anthracis
J.Biol.Chem., 284, 2009
|
|
1Z1D
 
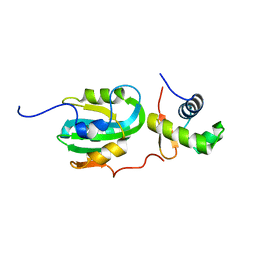 | | Structural Model for the interaction between RPA32 C-terminal domain and SV40 T antigen origin binding domain. | | Descriptor: | Large T antigen, Replication protein A 32 kDa subunit | | Authors: | Arunkumar, A.I, Klimovich, V, Jiang, X, Ott, R.D, Mizoue, L, Fanning, E, Chazin, W.J. | | Deposit date: | 2005-03-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1NLN
 
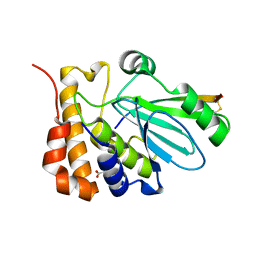 | | CRYSTAL STRUCTURE OF HUMAN ADENOVIRUS 2 PROTEINASE WITH ITS 11 AMINO ACID COFACTOR AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, Adenain, PVIC | | Authors: | McGrath, W.J, Ding, J, Sweet, R.M, Mangel, W.F. | | Deposit date: | 2003-01-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold
Biochim.Biophys.Acta, 1648, 2003
|
|
5ZLC
 
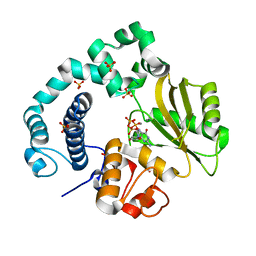 | | Binary complex of human DNA Polymerase Mu with MndGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA-directed DNA/RNA polymerase mu, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-03-27 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
3DV2
 
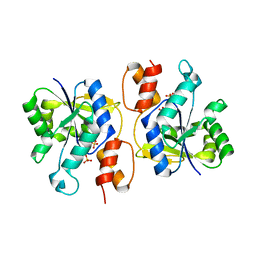 | | Crystal Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis | | Descriptor: | Nicotinate (Nicotinamide) nucleotide adenylyltransferase, SULFATE ION | | Authors: | Lu, S, Smith, C.D, Yang, Z, Pruett, P.S, Nagy, L, McCombs, D.P, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
3CJJ
 
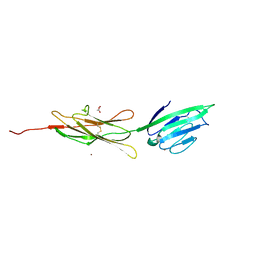 | | Crystal structure of human rage ligand-binding domain | | Descriptor: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | Authors: | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
5GSX
 
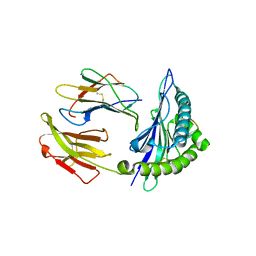 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-2 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5G2N
 
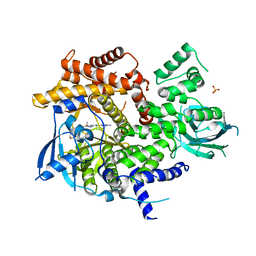 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
3GAE
 
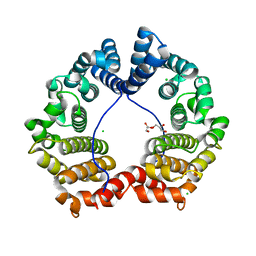 | | Crystal Structure of PUL | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DOA1 | | Authors: | Zhao, G, Schindelin, H, Lennarz, W.J. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8ITA
 
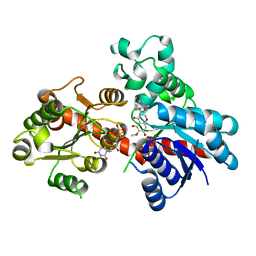 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT complexed with UDP and tectorigenin | | Descriptor: | 3-(4-hydroxyphenyl)-6-methoxy-5,7-bis(oxidanyl)chromen-4-one, Bc7OUGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
8JBG
 
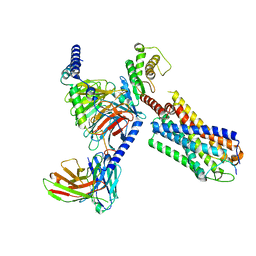 | | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBH
 
 | | Substance P bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
2COL
 
 | | Crystal structure analysis of CyaA/C-Cam with Pyrophosphate | | Descriptor: | Bifunctional hemolysin-adenylate cyclase, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Tang, W.J, Shen, Y. | | Deposit date: | 2005-05-18 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin
Embo J., 24, 2005
|
|
7XTY
 
 | | Crystal Structure of the second PDZ domain from human PTPN13 in complex with APC peptide | | Descriptor: | APC-peptide, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Jing, L.Q, Sun, X.N, Ma, W.H, Zhou, W.J, Wu, D.L. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting PTPN13 with 11 amino acid peptides of C-terminal APC prevents immune evasion of colorectal cancer
to be published
|
|
7Y5M
 
 | |
2BEP
 
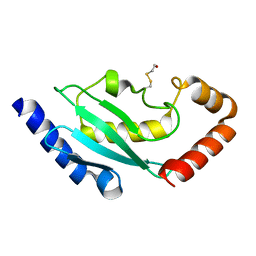 | | Crystal structure of ubiquitin conjugating enzyme E2-25K | | Descriptor: | BETA-MERCAPTOETHANOL, UBIQUITIN-CONJUGATING ENZYME E2-25 KDA | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
7YLK
 
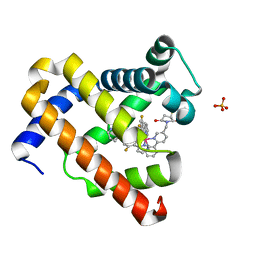 | | Myoglobin containing Ir complex | | Descriptor: | Myoglobin, SULFATE ION, delta-{1-([2,2'-bipyridin]-5-ylmethyl)pyrrolidine-2,5-dione}bis[2-(2,4-difluorophenyl)pyridine)]iridium(III), ... | | Authors: | Lee, J.H, Song, W.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Photocatalytic C-O Coupling Enzymes That Operate via Intramolecular Electron Transfer.
J.Am.Chem.Soc., 145, 2023
|
|
7YPL
 
 | |
2BF8
 
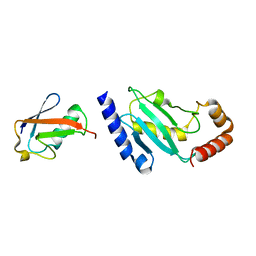 | | Crystal structure of SUMO modified ubiquitin conjugating enzyme E2- 25K | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-25 KDA, UBIQUITIN-LIKE PROTEIN SMT3C | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
7XQT
 
 | | The structure of FLA-K*00701/KP-FECV-11 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQS
 
 | | The structure of FLA-K*00701/KP-CoV-9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQU
 
 | | The structure of FLA-E*00301/EM-FECV-10 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide from Nucleoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
2B0Y
 
 | | Solution Structure of a peptide mimetic of the fourth cytoplasmic loop of the G-protein coupled CB1 cannabinoid receptor | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Grace, C.R, Cowsik, S.M, Shim, J.Y, Welsh, W.J, Howlett, A.C. | | Deposit date: | 2005-09-15 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unique helical conformation of the fourth cytoplasmic loop of the CB1 cannabinoid receptor in a negatively charged environment.
J.Struct.Biol., 159, 2007
|
|
