3RHY
 
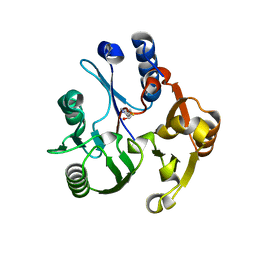 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with 4-chloro-2-hydroxymethylpyridine | | Descriptor: | (4-chloropyridin-2-yl)methanol, N(G),N(G)-dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Johnson, C.M, Ke, Z, Yoon, D.-W, Linsky, T.W, Guo, H, Fast, W, Robertus, J.D. | | Deposit date: | 2011-04-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | On the mechanism of dimethylarginine dimethylaminohydrolase inactivation by 4-halopyridines.
J.Am.Chem.Soc., 133, 2011
|
|
6VCL
 
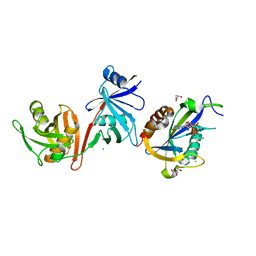 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCR
 
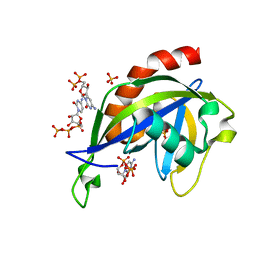 | | Crystal structure of E.coli RppH in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, PYROPHOSPHATE, RNA pyrophosphohydrolase, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VEK
 
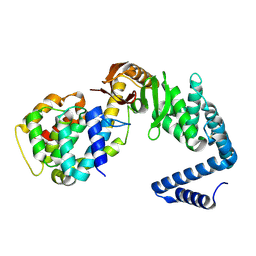 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length | | Descriptor: | contact-dependent immunity protein CdiI, contact-dependent toxin CdiA | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length
To Be Published
|
|
3R0H
 
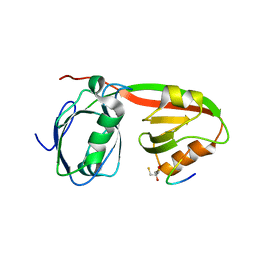 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
6VIZ
 
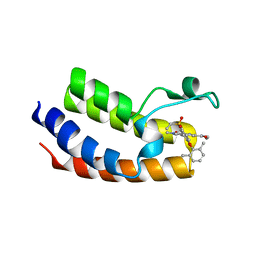 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VRO
 
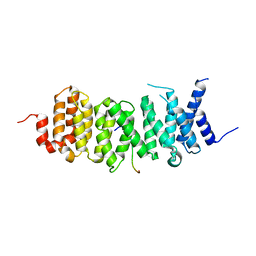 | | The structure of the PP2A B56 subunit AIM1 complex | | Descriptor: | Beta/gamma crystallin domain-containing protein 1, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2020-02-08 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment.
Elife, 9, 2020
|
|
1YGE
 
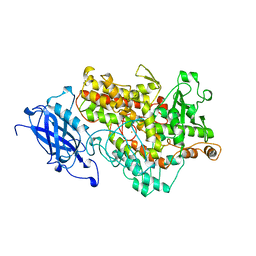 | | LIPOXYGENASE-1 (SOYBEAN) AT 100K | | Descriptor: | FE (III) ION, LIPOXYGENASE-1 | | Authors: | Minor, W, Steczko, J, Stec, B, Otwinowski, Z, Bolin, J.T, Walter, R, Axelrod, B. | | Deposit date: | 1996-06-04 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution.
Biochemistry, 35, 1996
|
|
6VQX
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7A0V
 
 | | Crystal structure of the 5-phosphatase domain of Synaptojanin1 in complex with a nanobody | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nanobody 13015, ... | | Authors: | Paesmans, J, Galicia, C, Martin, E, Versees, W. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations.
Elife, 9, 2020
|
|
6ZQU
 
 | | Cryo-EM structure of mature Dengue virus 2 at 3.1 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
7B4M
 
 | |
7B4L
 
 | |
6VIY
 
 | | BRD2_Bromodomain2 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 2 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6W8S
 
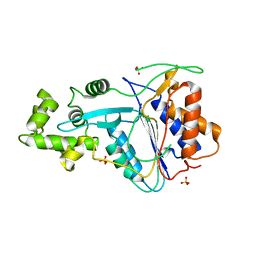 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
7B2P
 
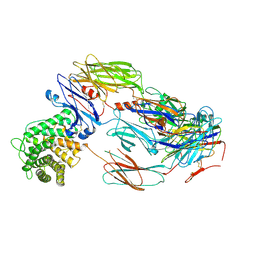 | | Cryo-EM structure of complement C4b in complex with nanobody B5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4 alpha chain, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
7B2M
 
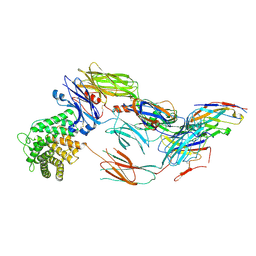 | | Cryo-EM structure of complement C4b in complex with nanobody E3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-C4b nanobody E3, Complement C4 alpha chain, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, van den Bos, R.M, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
7B2Q
 
 | | Cryo-EM structure of complement C4b in complex with nanobody B12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-C4b nanobody B12, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
3S69
 
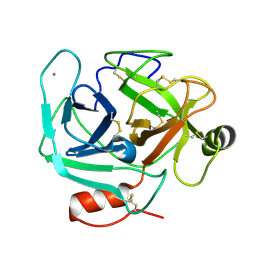 | | Crystal structure of saxthrombin | | Descriptor: | CALCIUM ION, Thrombin-like enzyme defibrase | | Authors: | Huang, K, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of saxthrombin, a thrombin-like enzyme from Gloydius saxatilis.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1XF0
 
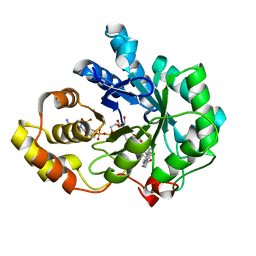 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 (AKR1C3) complexed with delta4-androstene-3,17-dione and NADP | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Qiu, W, Zhou, M, Labrie, F, Lin, S.-X. | | Deposit date: | 2004-09-13 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the multispecific 17beta-hydroxysteroid dehydrogenase type 5: critical androgen regulation in human peripheral tissues
Mol.Endocrinol., 18, 2004
|
|
7B7A
 
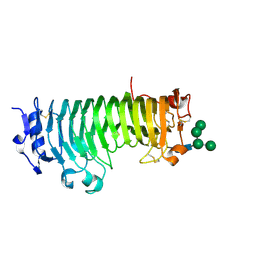 | | ENDO-POLYGALACTURONASE FROM ARABIDOPSIS THALIANA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pectin lyase-like superfamily protein, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Safran, J, Tabi, W, Habrylo, O, Bouckaert, J, Lefebvre, V, Senechal, F, Pelloux, J. | | Deposit date: | 2020-12-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Plant polygalacturonase structures specify enzyme dynamics and processivities to fine-tune cell wall pectins.
Plant Cell, 2023
|
|
7B8B
 
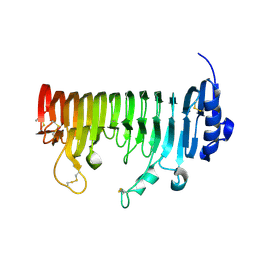 | | ADPG2 - ENDOPOLYGALACTURONASE FROM ARABIDOPSIS THALIANA | | Descriptor: | PHOSPHATE ION, Polygalacturonase ADPG2 | | Authors: | Safran, J, Tabi, W, Habrylo, O, Bouckaert, J, Lefebvre, V, Senechal, F, Pelloux, J. | | Deposit date: | 2020-12-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Plant polygalacturonase structures specify enzyme dynamics and processivities to fine-tune cell wall pectins.
Plant Cell, 2023
|
|
6W5D
 
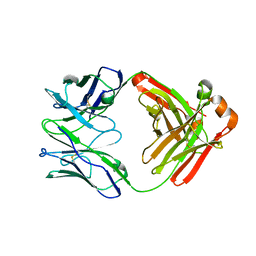 | | Crystal Structure of Fab RSB1 | | Descriptor: | RSB1 Fab Heavy Chain, RSB1 Fab Light Chain | | Authors: | Harshbarger, W, Chandramouli, S, Malito, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Convergent structural features of respiratory syncytial virus neutralizing antibodies and plasticity of the site V epitope on prefusion F.
Plos Pathog., 16, 2020
|
|
7AAZ
 
 | | Crystal structure of MerTK in complex with a type 1.5 aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-~{N}-[(1~{S},2~{S})-2-[[4-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]methoxy]cyclopentyl]-5-(1-methylpyrazol-4-yl)pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
6W8T
 
 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
