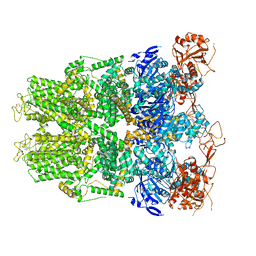
6OFB
 
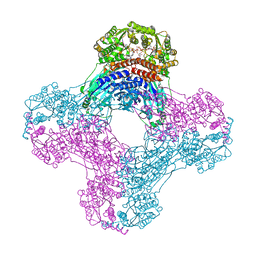 | | Crystal structure of human glutamine-dependent NAD+ synthetase complexed with NaAD+, AMP, pyrophosphate, and Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T.I, Gerratana, B. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Different ways to transport ammonia in human and Mycobacterium tuberculosis NAD+synthetases.
Nat Commun, 11, 2020
|
|
6DRK
 
 | | Structure of TRPM2 ion channel receptor by single particle electron cryo-microscopy, Apo state | | Descriptor: | Transient receptor potential cation channel, subfamily M, member 2 | | Authors: | Du, J, Lu, W, Huang, Y, Winkler, P, Sun, W. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium.
Nature, 562, 2018
|
|
6OM0
 
 | |
6O8A
 
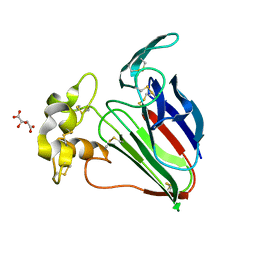 | | Thaumatin native-SAD structure determined at 5 keV from microcrystals | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Guo, G, Zhu, P, Fuchs, M.R, Shi, W, Andi, B, Gao, Y, Hendrickson, W.A, McSweeney, S, Liu, Q. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-08 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synchrotron microcrystal native-SAD phasing at a low energy.
Iucrj, 6, 2019
|
|
6OC9
 
 | | S8 phosphorylated beta amyloid 40 fibrils | | Descriptor: | Amyloid-beta precursor protein, PHOSPHONATE | | Authors: | Qiang, W, Hu, Z.W. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Molecular structure of an N-terminal phosphorylated beta-amyloid fibril.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OES
 
 | | Cryo-EM structure of mouse RAG1/2 STC complex (without NBD domain) | | Descriptor: | CALCIUM ION, DNA (34-MER), DNA (35-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OLE
 
 | |
6DJY
 
 | | Fako virus | | Descriptor: | Clamp protein, Major capsid protein, Turret protein | | Authors: | Kaelber, J.T, Jiang, W, Weaver, S.C, Auguste, A.J, Chiu, W. | | Deposit date: | 2018-05-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The polymerase organization of Fako virus
To be Published
|
|
6O47
 
 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | Descriptor: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
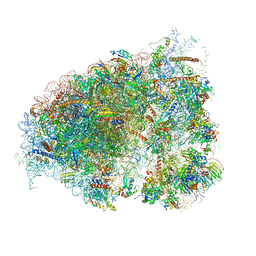
6O7K
 
 | | 30S initiation complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Frank, J, Gonzalez Jr, R.L, kaledhonkar, S, Fu, Z, Caban, K, Li, W, Chen, B, Sun, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-29 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Late steps in bacterial translation initiation visualized using time-resolved cryo-EM.
Nature, 570, 2019
|
|
6OEM
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | Descriptor: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6DVR
 
 | | Crystal structure of human CARM1 with (R)-SKI-72 | | Descriptor: | (2R,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-methoxyphenyl)ethyl]hexanamide (non-preferred name), 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Zeng, H, Hutchinson, A, Seitova, A, Luo, M, Cai, X.C, Ke, W, Wang, J, Shi, C, Zheng, W, Lee, J.P, Ibanez, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human CARM1 with (R)-SKI-72
to be published
|
|
4DQP
 
 | | Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and ddCTP (paired with dG of template) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*C*AP*TP*GP*GP*GP*AP*GP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DOC))-3'), ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-02-16 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
4DSE
 
 | | Ternary complex of Bacillus DNA Polymerase I Large Fragment F710Y, DNA duplex, and rCTP (paired with dG of template) in presence of Mg2+ | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*GP*GP*GP*AP*GP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DOC))-3'), ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
4E0D
 
 | | Binary complex of Bacillus DNA Polymerase I Large Fragment E658A and duplex DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
3C38
 
 | |
3C3E
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and GDP. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
3BXN
 
 | | The high resolution crystal structure of HLA-B*1402 complexed with a Cathepsin A signal sequence peptide, pCatA | | Descriptor: | Cathepsin A signal sequence octapeptide, GLYCEROL, HLA-B*1402 extracellular domain, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2008-01-14 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
3BPB
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase H162G adduct with S-methyl-L-thiocitrulline | | Descriptor: | N~5~-[(E)-imino(methylsulfanyl)methyl]-L-ornithine, dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Linsky, T.W, Stone, E.M, Fast, W, Robertus, J.D. | | Deposit date: | 2007-12-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Promiscuous partitioning of a covalent intermediate common in the pentein superfamily.
Chem.Biol., 15, 2008
|
|
3BQJ
 
 | | VA387 polypeptide | | Descriptor: | va387 polypeptide | | Authors: | Bu, W, Mamedova, A, Tan, M, Jiang, J, Hegde, R. | | Deposit date: | 2007-12-20 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
3BZP
 
 | | Crystal structural of the mutated N262A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZR
 
 | | Crystal structure of EscU C-terminal domain with N262D mutation, Space group P 41 21 2 | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZY
 
 | | Crystal structure of the mutated Y316D EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3C4J
 
 | | ABC protein ArtP in complex with ATP-gamma-S | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-30 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
3C5C
 
 | | Crystal structure of human Ras-like, family 12 protein in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-like protein 12, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human Ras-like, family 12 protein in complex with GDP.
To be Published
|
|
