2VE1
 
 | | Isopenicillin N synthase with substrate analogue AsMCOV (oxygen exposed 1min 20bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R,2S)-1-({[(1R)-1-carboxy-2-methylpropyl]oxy}carbonyl)-2-sulfanylpropyl]-6-oxo-L-lysine, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-10-15 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with a Sterically Demanding Depsipeptide Substrate Analogue.
Chembiochem, 10, 2009
|
|
2VCI
 
 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
2VCJ
 
 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-(5-chloro-2,4-dihydroxyphenyl)-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]isoxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
4ZMK
 
 | |
1DP8
 
 | |
4Z3S
 
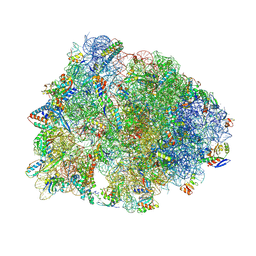 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic A201A, mRNA and three tRNAs in the A, P and E sites at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 3'-deoxy-3'-{[(2E)-3-(4-{[(4Z)-6-O-(6-deoxy-3,4-di-O-methyl-alpha-D-mannopyranosyl)-5-O-methyl-alpha-D-threo-hex-4-enofuranosyl]oxy}phenyl)-2-methylprop-2-enoyl]amino}-N,N-dimethyladenosine, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-03-31 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
7MBR
 
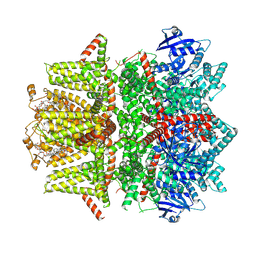 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (apo state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4ZFL
 
 | |
7MBS
 
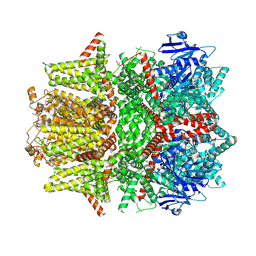 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (open state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBT
 
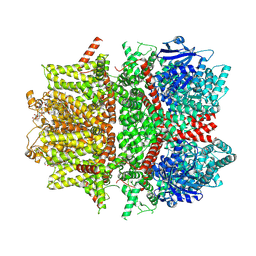 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (low calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBQ
 
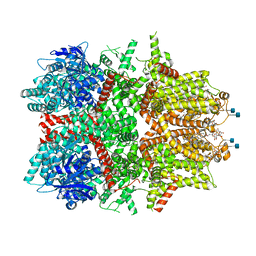 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MOY
 
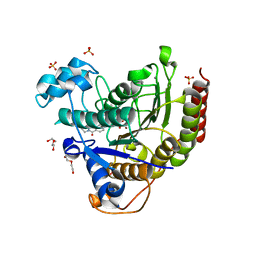 | | Structure of HDAC2 in complex with an inhibitor (compound 19) | | Descriptor: | (1S)-6-ethyl-N-{(1S)-1-[5-(2-ethyl-1-oxo-1,2-dihydroisoquinolin-6-yl)-1H-imidazol-2-yl]-7,7-dihydroxynonyl}-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MBU
 
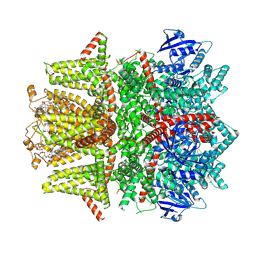 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (high calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBV
 
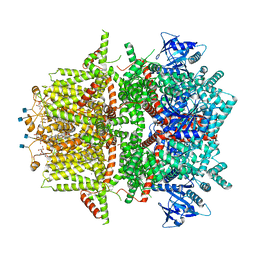 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium and 0.5 mM NDNA | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2WFD
 
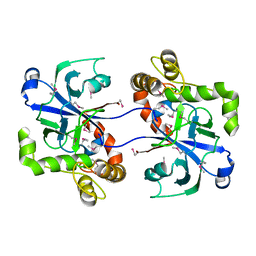 | | Structure of the human cytosolic leucyl-tRNA synthetase editing domain | | Descriptor: | LEUCYL-TRNA SYNTHETASE, CYTOPLASMIC | | Authors: | Seiradake, E, Mao, W, Hernandez, V, Baker, S.J, Plattner, J.J, Alley, M.R.K, Cusack, S. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of the Human and Fungal Cytosolic Leucyl-tRNA Synthetase Editing Domains: A Structural Basis for the Rational Design of Antifungal Benzoxaboroles.
J.Mol.Biol., 390, 2009
|
|
7MOT
 
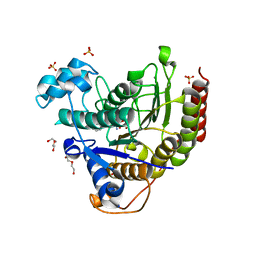 | | Structure of HDAC2 in complex with an inhibitor (compound 9) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOS
 
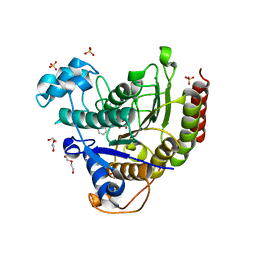 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 4) | | Descriptor: | (3S,18S,20aR)-18-(6,6-dihydroxyoctyl)-1,5,6,7,8,18,19,20a-octahydro-4H-14,17-epiminoazeto[1,2-g][1,7,10,13]benzoxatriazacycloheptadecin-20(2H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOX
 
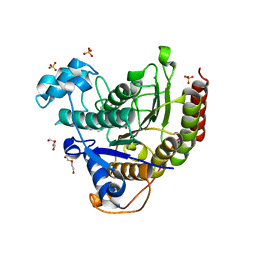 | | Structure of HDAC2 in complex with an inhibitor (compound 14) | | Descriptor: | (1S)-N-[(1S)-7,7-dihydroxy-1-{4-[(1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-6-yl]-1H-imidazol-2-yl}nonyl]-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOZ
 
 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 25) | | Descriptor: | (1R,3S,6S,18R,27R)-6-(6,6-dihydroxyoctyl)-5,8,18,27,34-pentaazahexacyclo[25.2.2.1~7,10~.1~11,15~.1~14,18~.0~1,3~]tetratriaconta-7,9,11(33),12,14,16-hexaene-4,32-dione (non-preferred name), CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
4YY8
 
 | | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum | | Descriptor: | GLYCEROL, Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum.
to be published
|
|
4ZMH
 
 | | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nocek, B, Cui, H, Wang, W, Savchenko, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biochemical and Structural Characterization of a Five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T.
J.Biol.Chem., 291, 2016
|
|
5A4N
 
 | |
2WFU
 
 | | Crystal structure of DILP5 variant DB | | Descriptor: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | Authors: | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | Deposit date: | 2009-04-15 | | Release date: | 2010-05-26 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
5A7T
 
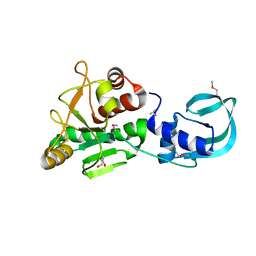 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.4 angstrom resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
2WGU
 
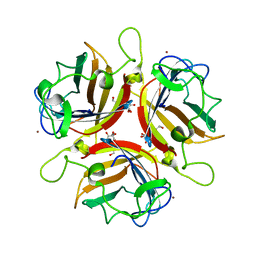 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N- methoxycarbonyl -3,5-dideoxy- D-glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-[(methoxycarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
