4V52
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8C3D
 
 | | Sulfonated Calpeptin is a promising drug candidate against SARS-CoV-2 infections | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CALCIUM ION, Cathepsin K | | Authors: | Loboda, J, Karnicar, K, Lindic, N, Usenik, A, Lieske, J, Meents, A, Guenther, S, Reinke, P.Y.A, Falke, S, Ewert, W, Turk, D. | | Deposit date: | 2022-12-23 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
2VPR
 
 | | Tet repressor class H in complex with 5a,6- anhydrotetracycline-Mg | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Schuldt, L, Palm, G, Hinrichs, W. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Tet Repressor Induction by Tetracycline: A Molecular Dynamics, Continuum Electrostatics, and Crystallographic Study
J.Mol.Biol., 378, 2008
|
|
5LY6
 
 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
4UXL
 
 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5EJC
 
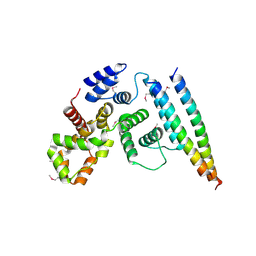 | | Crystal structural of the TSC1-TBC1D7 complex | | Descriptor: | Hamartin, TBC1 domain family member 7 | | Authors: | Wang, Z, Qin, J, Gong, W, Xu, W. | | Deposit date: | 2015-11-01 | | Release date: | 2016-03-02 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of the Interaction between Tuberous Sclerosis Complex 1 (TSC1) and Tre2-Bub2-Cdc16 Domain Family Member 7 (TBC1D7).
J.Biol.Chem., 291, 2016
|
|
6Z1V
 
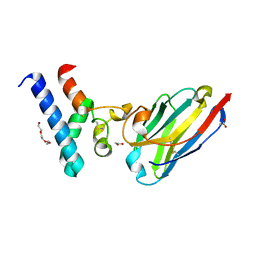 | | Structure of the EC2 domain of CD9 in complex with nanobody 4E8 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CD9 antigen, ... | | Authors: | Oosterheert, W, Pearce, N.M, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6Z20
 
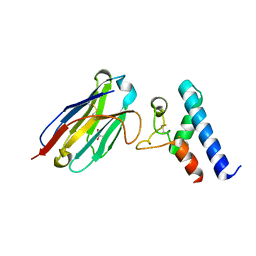 | | Structure of the EC2 domain of CD9 in complex with nanobody 4C8 | | Descriptor: | CD9 antigen, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oosterheert, W, Manshande, J, Pearce, N.M, Lutz, M, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
5M04
 
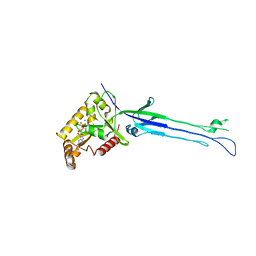 | | Structure of ObgE from Escherichia coli | | Descriptor: | GTPase ObgE/CgtA, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gkekas, S, Singh, R.K, Versees, W. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J. Biol. Chem., 292, 2017
|
|
5LNF
 
 | | Solution NMR structure of farnesylated PEX19, C-terminal domain | | Descriptor: | FARNESYL, Peroxisomal biogenesis factor 19 | | Authors: | Emmanouilidis, L, Schuetz, U, Tripsianes, K, Madl, T, Radke, J, Rucktaeschel, R, Wilmanns, M, Schliebs, W, Erdmann, R, Sattler, M. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-15 | | Last modified: | 2019-09-11 | | Method: | SOLUTION NMR | | Cite: | Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19.
Nat Commun, 8, 2017
|
|
2IRW
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Ether Inhibitor | | Descriptor: | (1S,3R,4S,5S,7S)-4-{[2-(4-METHOXYPHENOXY)-2-METHYLPROPANOYL]AMINO}ADAMANTANE-1-CARBOXAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Longenecker, K.L, Patel, J.R, Russell, J, Qin, W. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of adamantane ethers as inhibitors of 11beta-HSD-1: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2ILT
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Sulfone Inhibitor | | Descriptor: | 2-(2-CHLORO-4-FLUOROPHENOXY)-2-METHYL-N-[(1R,2S,3S,5S,7S)-5-(METHYLSULFONYL)-2-ADAMANTYL]PROPANAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Longenecker, K.L, Sorensen, B, Judge, R, Qin, W, Link, J.T. | | Deposit date: | 2006-10-03 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adamantane sulfone and sulfonamide 11-beta-HSD1 Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3EQY
 
 | |
3EQS
 
 | |
7NXF
 
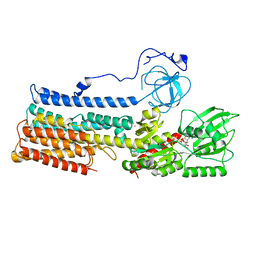 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
7NY1
 
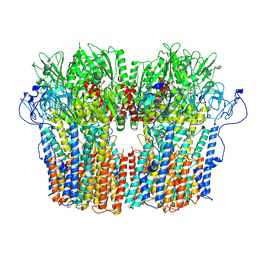 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
8HR6
 
 | |
8HPE
 
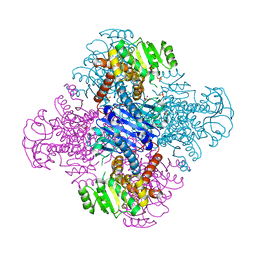 | | Crystal structure of Leucine dehydrogenase | | Descriptor: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | Authors: | Li, X, Song, W. | | Deposit date: | 2022-12-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
1K5N
 
 | | HLA-B*2709 BOUND TO NONA-PEPTIDE M9 | | Descriptor: | GLYCEROL, beta-2-microglobulin, light chain, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | HLA-B27 Subtypes Differentially Associated with Disease Exhibit Subtle Structural Alterations
J.Biol.Chem., 277, 2002
|
|
2W6C
 
 | | ACHE IN COMPLEX WITH A BIS-(-)-NOR-MEPTAZINOL DERIVATIVE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3R)-3-ethyl-1-{9-[(3S)-3-ethyl-3-(3-hydroxyphenyl)azepan-1-yl]nonyl}azepan-3-yl]phenol, ... | | Authors: | Paz, A, Xie, Q, Greenblatt, H.M, Fu, W, Tang, Y, Silman, I, Qiu, Z, Sussman, J.L. | | Deposit date: | 2008-12-18 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The Crystal Structure of a Complex of Acetylcholinesterase with a Bis-(-)-Nor-Meptazinol Derivative Reveals Disruption of the Catalytic Triad.
J.Med.Chem., 52, 2009
|
|
1KFX
 
 | | Crystal Structure of Human m-Calpain Form I | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4V54
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
4V0I
 
 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4W5U
 
 | |
