1SZ3
 
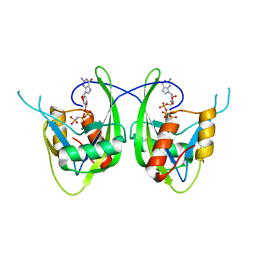 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEXED WITH GNP AND MG+2 | | Descriptor: | MAGNESIUM ION, MutT/nudix family protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-04-02 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1LNL
 
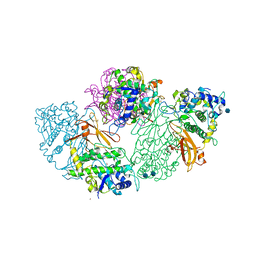 | | Structure of deoxygenated hemocyanin from Rapana thomasiana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, hemocyanin | | Authors: | Perbandt, M, Guthoehrlein, E.W, Rypniewski, W, Idakieva, K, Stoeva, S, Voelter, W, Genov, N, Betzel, C. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Biochemistry, 42, 2003
|
|
1SU2
 
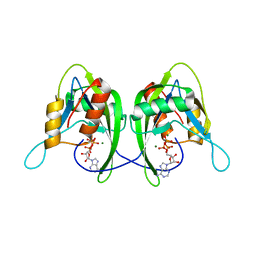 | | CRYSTAL STRUCTURE OF THE NUDIX HYDROLASE DR1025 IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
2RGM
 
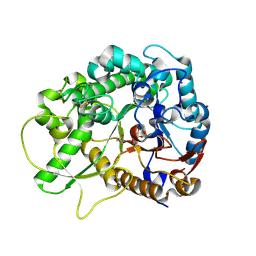 | | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2007-10-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Rice BGlu1 beta-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation
J.Mol.Biol., 377, 2008
|
|
2F5T
 
 | | Crystal Structure of the sugar binding domain of the archaeal transcriptional regulator TrmB | | Descriptor: | IMIDAZOLE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, archaeal transcriptional regulator TrmB | | Authors: | Krug, M, Lee, S.J, Diederichs, K, Boos, W, Welte, W. | | Deposit date: | 2005-11-27 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Sugar Binding Domain of the Archaeal Transcriptional Regulator TrmB
J.Biol.Chem., 281, 2006
|
|
1L8Z
 
 | | Solution structure of HMG box 5 in human upstream binding factor | | Descriptor: | upstream binding factor 1 | | Authors: | Yang, W, Xu, Y, Wu, J, Zeng, W, Shi, Y. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor
Biochemistry, 42, 2003
|
|
2ROW
 
 | | The C1 domain of ROCK II | | Descriptor: | Rho-associated protein kinase 2, ZINC ION | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C1 domain of ROCK II
To be Published
|
|
4HJX
 
 | | Crystal structure of F2YRS complexed with F2Y | | Descriptor: | 3,5-difluoro-L-tyrosine, Tyrosine-tRNA ligase | | Authors: | Wang, J, Tian, C, Gong, W, Li, F, Shi, P, Li, J, Ding, W. | | Deposit date: | 2012-10-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A genetically encoded 19F NMR probe for tyrosine phosphorylation.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1LG9
 
 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-15 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
3IB3
 
 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
1LGF
 
 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-15 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
2Q58
 
 | | Cryptosporidium parvum putative polyprenyl pyrophosphate synthase (cgd4_2550) in complex with zoledronate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, ZOLEDRONIC ACID | | Authors: | Chruszcz, M, Artz, J, Zheng, H, Dong, A, Dunford, J, Lew, J, Zhao, Y, Kozieradski, I, Kavanaugh, K.L, Opperman, U, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Bochkarev, A, Hui, R, Minor, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Targeting a uniquely nonspecific prenyl synthase with bisphosphonates to combat cryptosporidiosis
Chem.Biol., 15, 2008
|
|
2K13
 
 | | Solution NMR Structure of the Leech Protein Saratin, a Novel Inhibitor of Haemostasis | | Descriptor: | Saratin | | Authors: | Gronwald, W, Bomke, J, Maurer, T, Wisotzki, B, Huber, F, Schumann, F, Kremer, W, Frech, M, Kalbitzer, H.R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the leech protein saratin and characterization of its binding to collagen
J.Mol.Biol., 381, 2008
|
|
1BTH
 
 | |
2PRK
 
 | |
3K9V
 
 | | Crystal structure of rat mitochondrial P450 24A1 S57D in complex with CHAPS | | Descriptor: | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Annalora, A.J, Goodin, D.B, Hong, W, Zhang, Q, Johnson, E.F, Stout, C.D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism.
J.Mol.Biol., 396, 2010
|
|
7T4J
 
 | | Crystal Structure of EGFR_D770_N771insNPG/V948R in complex with TAK-788 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Epidermal growth factor receptor, ... | | Authors: | Skene, R.J, Lane, W, Hu, Y. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7T4I
 
 | | Crystal Structure of wild type EGFR in complex with TAK-788 | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Skene, R.J, Lane, W. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
5KU3
 
 | | BRD4 bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, Bromodomain-containing protein 4, ... | | Authors: | Murray, J.M, Huang, W. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
3TPI
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, ISOLEUCINE, ... | | Authors: | Huber, R, Bode, W, Deisenhofer, J, Schwager, P. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
7V3T
 
 | | Solution structure of thrombin binding aptamer G-quadruplex bound a self-adaptive small molecule with rotated ligands | | Descriptor: | 11,13-bis(fluoranyl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-7$l^{4}-aza-8$l^{4}-platinatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene, TBA G4 DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Liu, W, Zhu, B.C, Mao, Z.W. | | Deposit date: | 2021-08-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound.
Chem Sci, 13, 2022
|
|
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone
To Be Published
|
|
5LY6
 
 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
