5L2T
 
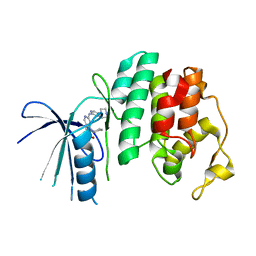 | | The X-ray co-crystal structure of human CDK6 and Ribociclib. | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-{[5-(piperazin-1-yl)pyridin-2-yl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Cyclin-dependent kinase 6 | | Authors: | Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance.
Mol.Cancer Ther., 15, 2016
|
|
6XAH
 
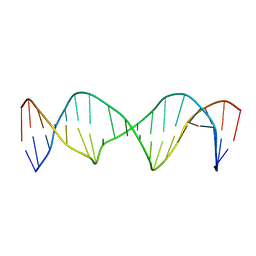 | | Structure of a Stable Interstrand DNA Crosslink Involving an dA Amino Group and an Abasic Site | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*GP*AP*AP*CP*(AAB)P*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*TP*CP*TP*AP*AP*GP*TP*TP*CP*AP*TP*CP*TP*A)-3') | | Authors: | Kellum Jr, A.H, Qiu, D, Voehler, M.W, Martin, W.J, Gates, K.S, Stone, M.P. | | Deposit date: | 2020-06-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable Interstrand DNA Cross-Link Involving a beta- N -Glycosyl Linkage Between an N 6 -dA Amino Group and an Abasic Site.
Biochemistry, 60, 2021
|
|
6XOS
 
 | |
6XOU
 
 | |
6XOV
 
 | |
6XOT
 
 | |
6YHL
 
 | |
6SY1
 
 | | Crystal structure of the human 2-oxoadipate dehydrogenase DHTKD1 (E1) | | Descriptor: | MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, ... | | Authors: | Bezerra, G.A, Foster, W, Shrestha, L, Pena, I.A, Coker, J, Kolker, S, Nicola, B.B, von Delft, F, Edwards, A, Arrowsmith, C, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism.
Iucrj, 7, 2020
|
|
6Y4F
 
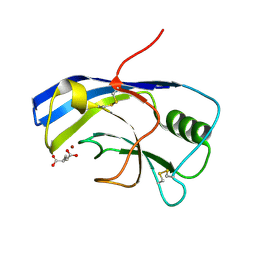 | | X-ray structure of the Zn-dependent receptor-binding domain of Proteus mirabilis MR/P fimbrial adhesin MrpH | | Descriptor: | Fimbrial adhesin, GLUTAMIC ACID, ZINC ION | | Authors: | Knight, S.D, Ubhayasekera, W, Jiang, W. | | Deposit date: | 2020-02-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MrpH, a new class of metal-binding adhesin, requires zinc to mediate biofilm formation.
Plos Pathog., 16, 2020
|
|
6Y4E
 
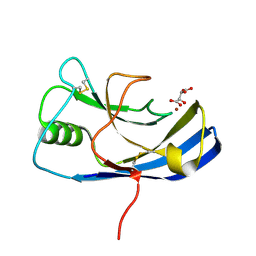 | | X-ray structure of the Zn-dependent receptor-binding domain of Proteus mirabilis MR/P fimbrial adhesin MrpH | | Descriptor: | Fimbrial adhesin, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Knight, S.D, Ubhayasekera, W, Jiang, W. | | Deposit date: | 2020-02-20 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | MrpH, a new class of metal-binding adhesin, requires zinc to mediate biofilm formation.
Plos Pathog., 16, 2020
|
|
5M5W
 
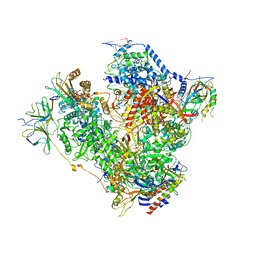 | | RNA Polymerase I open complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Hoffmann, N.A, Jakobi, A.J, Wetzel, R, Hagen, W.J.H, Sachse, C, Muller, C.W. | | Deposit date: | 2016-10-23 | | Release date: | 2016-12-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular Structures of Transcribing RNA Polymerase I.
Mol. Cell, 64, 2016
|
|
5M5Y
 
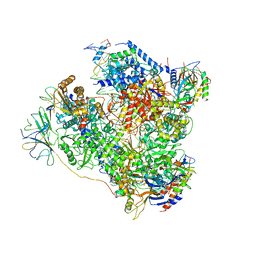 | | RNA Polymerase I elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Hoffmann, N.A, Jakobi, A.J, Wetzel, R, Hagen, W.J.H, Sachse, C, Muller, C.W. | | Deposit date: | 2016-10-23 | | Release date: | 2016-12-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular Structures of Transcribing RNA Polymerase I.
Mol. Cell, 64, 2016
|
|
4QYY
 
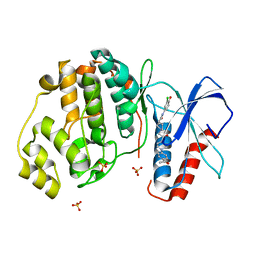 | | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase State | | Descriptor: | (3R)-1-{2-[4-(4-acetylphenyl)piperazin-1-yl]-2-oxoethyl}-N-(3-chloro-4-hydroxyphenyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Deng, Y, Shipps, G.W, Cooper, A, English, J.M, Annis, D.A, Carr, D, Nan, Y, Wang, T, Zhu, Y.H, Chuang, C, Dayananth, P, Hruza, A.W, Xiao, L, Jin, W, Kirschmeier, P, Windsor, W.T, Samatar, A.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase.
J.Med.Chem., 57, 2014
|
|
2CK3
 
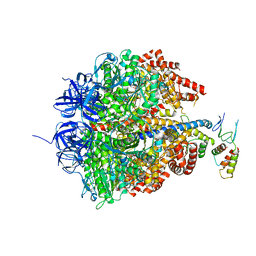 | | Azide inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How Azide Inhibits ATP Hydrolysis by the F-Atpases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5M5X
 
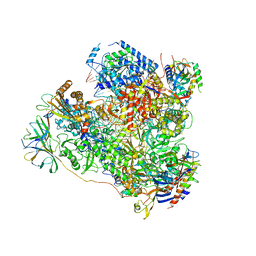 | | RNA Polymerase I elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Hoffmann, N.A, Jakobi, A.J, Wetzel, R, Hagen, W.J.H, Sachse, C, Muller, C.W. | | Deposit date: | 2016-10-23 | | Release date: | 2016-12-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular Structures of Transcribing RNA Polymerase I.
Mol. Cell, 64, 2016
|
|
4R0V
 
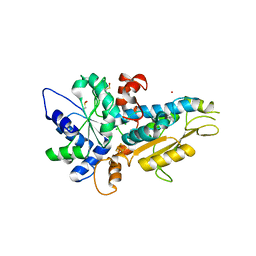 | | [FeFe]-hydrogenase Oxygen Inactivation is Initiated by the Modification and Degradation of the H cluster 2Fe Subcluster | | Descriptor: | ARSENIC, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Swanson, S.D, Ratzloff, M.W, Mulder, D.W, Artz, J.H, Ghose, S, Hoffman, A, White, S, Zadvornyy, O.A, Broderick, J.B, Bothner, B, King, P.W, Peters, J.W. | | Deposit date: | 2014-08-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | [FeFe]-Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster.
J.Am.Chem.Soc., 137, 2015
|
|
2Y6W
 
 | | Structure of a Bcl-w dimer | | Descriptor: | BCL-2-LIKE PROTEIN 2, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Lee, E.F, Evangelista, M, Pettikiriarachchi, A, Dogovski, C, Perugini, M.A, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2011-01-27 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Bcl-W Domain-Swapped Dimer: Implications for the Function of Bcl-2 Family Proteins.
Structure, 19, 2011
|
|
6Z2K
 
 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
8BOT
 
 | |
8K1T
 
 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Nat Commun, 15, 2024
|
|
1ZD0
 
 | | Crystal structure of Pfu-542154 conserved hypothetical protein | | Descriptor: | MAGNESIUM ION, METHANOL, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J.E, Liu, Z.J, Horanyi, P.S, Florence, Q.J.T, Tempel, W, Zhou, W, Chen, L, Lee, D, Nguyen, J, Chang, S.H, Bereton, P, Izumi, M, Jenny Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-13 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pfu-542154 conserved hypothetical protein
To be Published
|
|
8K1U
 
 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Nat Commun, 15, 2024
|
|
8K1S
 
 | | Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Nat Commun, 15, 2024
|
|
1YD7
 
 | | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha, UNKNOWN ATOM OR ION | | Authors: | Horanyi, P, Florence, Q, Zhou, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus
To be published
|
|
2HBS
 
 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF DEOXYHEMOGLOBIN S | | Descriptor: | HEMOGLOBIN S (DEOXY), ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Harrington, D.J, Adachi, K, Royer Junior, W.E. | | Deposit date: | 1997-05-06 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The high resolution crystal structure of deoxyhemoglobin S.
J.Mol.Biol., 272, 1997
|
|
