6EV2
 
 | | Crystal structure of antibody against schizophyllan in complex with laminarihexaose | | Descriptor: | Heavy chain, Light chain, beta-D-glucopyranose, ... | | Authors: | Sung, K.H, Josewski, J, Duebel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
1IIG
 
 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHONOPROPIONATE | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
6UON
 
 | |
6UP2
 
 | |
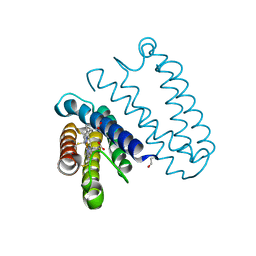
5JVE
 
 | |
5JW3
 
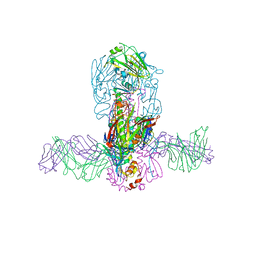 | | Structure of MEDI8852 Fab Fragment in Complex with H7 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Collins, P.J, Neu, U, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
3GNZ
 
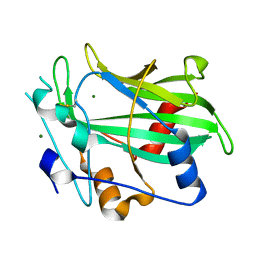 | | Toxin fold for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8UCU
 
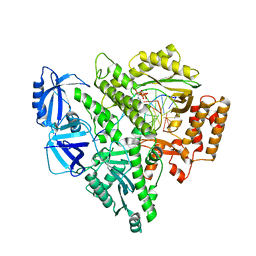 | | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2DCO
 
 | |
6PE4
 
 | | Yeast Vo motor in complex with 1 VopQ molecule | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PF4
 
 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)phenyl)acetic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)phenyl]acetic acid | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
8DTQ
 
 | | Crystal Structure of Staphylococcus aureus pSK41 Cop | | Descriptor: | CHLORIDE ION, Helix-turn-helix domain, SODIUM ION | | Authors: | Walton, W.G, Eakes, T.C, Redinbo, M.R, McLaughlin, K.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | pSK41/pGO1-family conjugative plasmids of Staphylococcus aureus encode a cryptic repressor of replication.
Plasmid, 128, 2023
|
|
6WMH
 
 | |
6WNA
 
 | | Next generation monomeric IgG4 Fc | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, Immunoglobulin heavy constant gamma 4, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WOL
 
 | | Next generation monomeric IgG4 Fc bound to neonatal Fc receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, IgG receptor FcRn large subunit p51, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6V7A
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2657 | | Descriptor: | Hdac6 protein, N-hydroxy-4-[(1-methyl-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)methyl]benzamide, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.0874176 Å) | | Cite: | Spiroindoline-Capped Selective HDAC6 Inhibitors: Design, Synthesis, Structural Analysis, and Biological Evaluation.
Acs Med.Chem.Lett., 11, 2020
|
|
8ULM
 
 | |
5K1S
 
 | | crystal structure of AibC | | Descriptor: | Oxidoreductase, zinc-binding dehydrogenase family, ZINC ION | | Authors: | Bock, T, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of AibC, a reductase involved in alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6PDN
 
 | | Human PIM1 bound to benzothiophene inhibitor 292 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxylic acid, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
6PDO
 
 | | Human PIM1 bound to benzothiophene inhibitor 354 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxamide, Peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
6X4J
 
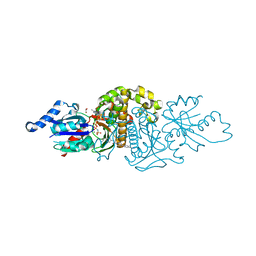 | | PANK3 complex structure with compound PZ-2863 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-{[4-(propan-2-yl)phenyl]acetyl}piperazin-1-yl)pyridine-3-carbonitrile, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LipE guided discovery of isopropylphenyl pyridazines as pantothenate kinase modulators.
Bioorg.Med.Chem., 52, 2021
|
|
5JOO
 
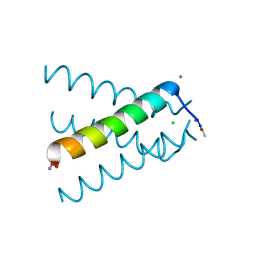 | | XFEL structure of influenza A M2 wild type TM domain at low pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2016-05-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3ELC
 
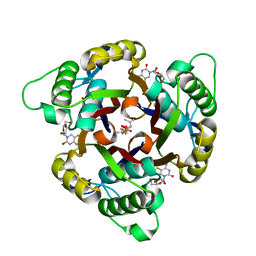 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with ligand | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-fluoro-pyrimidin-2-one, GERANYL DIPHOSPHATE, ... | | Authors: | Hunter, W.N, Ramsden, N.L, Ulaganathan, V. | | Deposit date: | 2008-09-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
6PDG
 
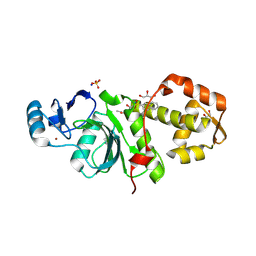 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 83 | | Descriptor: | 5-ethoxy-2-fluoro-3-methyl-N'-[(naphthalen-2-yl)sulfonyl]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6PDM
 
 | | Crystal structure of Human Protein Arginine Methyltransferase 9 (PRMT9) | | Descriptor: | Protein arginine N-methyltransferase 9, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Tempel, W, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Human Protein Arginine Methyltransferase 9 (PRMT9)
To Be Published
|
|
