1GM1
 
 | | Second PDZ Domain (PDZ2) of PTP-BL | | Descriptor: | PROTEIN TYROSINE PHOSPHATASE | | Authors: | Walma, T, Tessari, M, Aelen, J, Schepens, J, Hendriks, W, Vuister, G.W. | | Deposit date: | 2001-09-06 | | Release date: | 2002-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and binding characteristics of the second PDZ domain of PTP-BL.
J. Mol. Biol., 316, 2002
|
|
6ZAY
 
 | | Crystal structure of Atg16L in complex with GDP-bound Rab33B | | Descriptor: | 1,2-ETHANEDIOL, Autophagy-related protein 16-1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pantoom, S, Wu, Y.W. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RAB33B recruits the ATG16L1 complex to the phagophore via a noncanonical RAB binding protein.
Autophagy, 17, 2021
|
|
1S9W
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope, SLLMWITQC, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
6BPA
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 3E9 | | Descriptor: | BROMIDE ION, Monoclonal antibody 3E9 Fab heavy chain, Monoclonal antibody 3E9 Fab light chain, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6ZH4
 
 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7BOY
 
 | |
7B9V
 
 | | Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound | | Descriptor: | 5' exon of UBC4 mRNA, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0054360.mRNA.1.CDS.1, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Nagai, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for conformational equilibrium of the catalytic spliceosome.
Mol.Cell, 81, 2021
|
|
7BOX
 
 | |
6R2H
 
 | | Crystal structure of Apo PinO from Porphyromonas gingivitis | | Descriptor: | GLYCEROL, HmuY protein | | Authors: | Antonyuk, S.V, Bielecki, M, Strange, R.W, Capper, M, Olczak, T, Olczak, M. | | Deposit date: | 2019-03-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Prevotella intermedia produces two proteins homologous to Porphyromonas gingivalis HmuY but with different heme coordination mode.
Biochem.J., 477, 2020
|
|
1R31
 
 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
1GK1
 
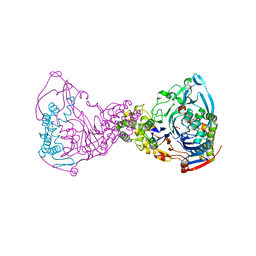 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1RH0
 
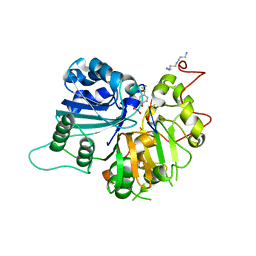 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine and trinucleotide GTT | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*GP*TP*T)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
8UVL
 
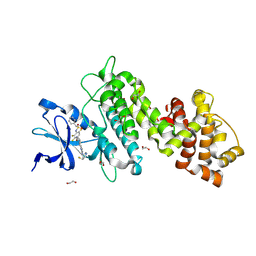 | | Crystal structure of selective IRE1a inhibitor 29 at the enzyme active site | | Descriptor: | 1,2-ETHANEDIOL, 1-phenyl-N-(2,3,6-trifluoro-4-{[(3M)-3-(2-{[(3R,5R)-5-fluoropiperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}phenyl)methanesulfonamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Kiefer, J.R, Wallweber, H.A, Braun, M.-G, Wei, W, Jiang, F, Wang, W, Rudolph, J, Ashkenazi, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Potent, Selective, and Orally Available IRE1 alpha Inhibitors Demonstrating Comparable PD Modulation to IRE1 Knockdown in a Multiple Myeloma Model.
J.Med.Chem., 67, 2024
|
|
6BFQ
 
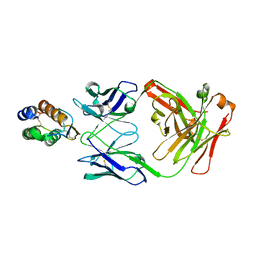 | | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies | | Descriptor: | Fab Heavy chain, Fab Light Chain, Granulocyte-macrophage colony-stimulating factor | | Authors: | Dhagat, U, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2017-10-26 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies suggests novel therapeutic opportunities.
MAbs, 10, 2018
|
|
6G4Q
 
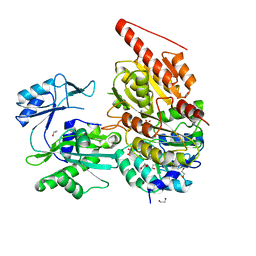 | | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2 | | Descriptor: | 1,2-ETHANEDIOL, Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial, ... | | Authors: | Bailey, H.J, Shrestha, L, Rembeza, E, Sorrell, F.J, Newman, J, Strain-Damerell, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2
To Be Published
|
|
3I2B
 
 | | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase | | Descriptor: | 1,2-ETHANEDIOL, 6-pyruvoyl tetrahydrobiopterin synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ugochukwu, E, Cocking, R, Pilka, E, Yue, W.W, Bray, J.E, Chaikuad, A, Krojer, T, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase
To be Published
|
|
3OY3
 
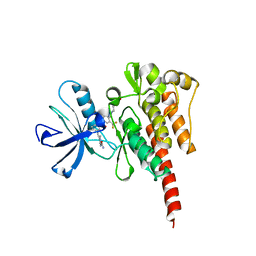 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
7U59
 
 | |
6ZH2
 
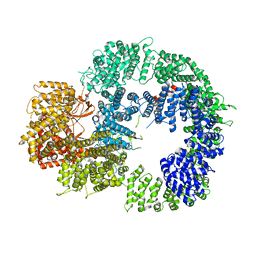 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1RGC
 
 | | THE COMPLEX BETWEEN RIBONUCLEASE T1 AND 3'-GUANYLIC ACID SUGGESTS GEOMETRY OF ENZYMATIC REACTION PATH. AN X-RAY STUDY | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Heydenreich, A, Koellner, G, Choe, H.W, Cordes, F, Kisker, C, Schindelin, H, Adamiak, R, Hahn, U, Saenger, W. | | Deposit date: | 1993-05-12 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex between ribonuclease T1 and 3'GMP suggests geometry of enzymic reaction path. An X-ray study.
Eur.J.Biochem., 218, 1993
|
|
4COO
 
 | | Crystal structure of human cystathionine beta-synthase (delta516-525) at 2.0 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, ... | | Authors: | McCorvie, T.J, Kopec, J, Vollamar, M, Strain-Damerell, C, Bushell, S, Bradley, A, Tallant, C, Kiyani, W, Froese, D.S, Carpenter, E.S, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-01-29 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
721P
 
 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
6ZH6
 
 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3UNO
 
 | |
7AE3
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 3 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
