7YMT
 
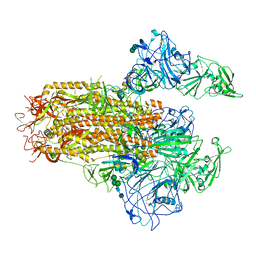 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (6.55 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YN0
 
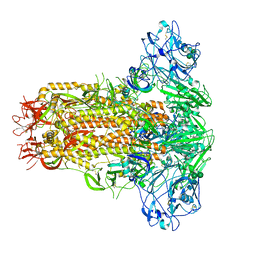 | | Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
6S3N
 
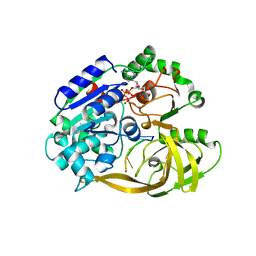 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP-VO4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.533 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
7YRS
 
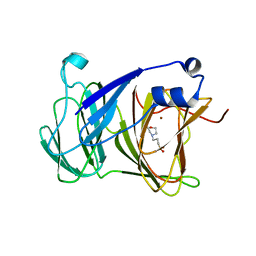 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7YPL
 
 | |
7YW1
 
 | | crystal structure of UBE2O | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme UBE2O | | Authors: | Fu, Z, Zhu, W, Huang, H. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | crystal structure of UBE2O
To Be Published
|
|
7SAN
 
 | | Crystal structure of human hypoxanthine guanine phosphoribzosyltransferase in complex with (4S,7S)-7-hydroxy-4-((guanin-9-yl)methyl)-2,5-dioxaheptan-1,7-diphosphonate | | Descriptor: | ({(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-[(2S)-2-hydroxy-2-phosphonoethoxy]propoxy}methyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Guddat, L.W, Keough, D.T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58155513 Å) | | Cite: | Stereo-Defined Acyclic Nucleoside Phosphonates are Selective and Potent Inhibitors of Parasite 6-Oxopurine Phosphoribosyltransferases.
J.Med.Chem., 65, 2022
|
|
6RLS
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (apo species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_apo | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
7YRR
 
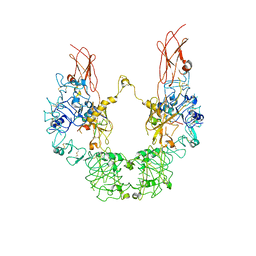 | | Cryo-EM structure of IGF1R with two IGF1 complex | | Descriptor: | Insulin-like growth factor 1 receptor, Isoform 3 of Insulin-like growth factor I | | Authors: | Xi, Z, Cang, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of IGF1R with two IGF1 complex at 4.3 angstroms resolution
To Be Published
|
|
7SBB
 
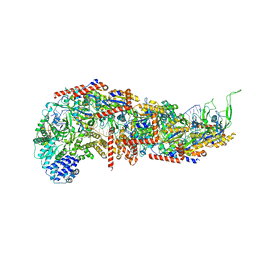 | |
7SBA
 
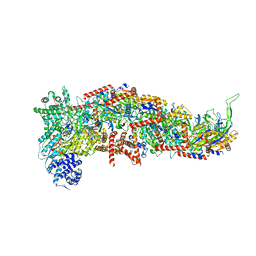 | |
7S9V
 
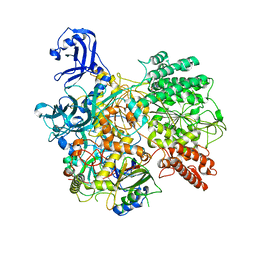 | | DrmAB:ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DrmA, DrmB | | Authors: | Bravo, J.P.K, Taylor, D.W, Brouns, S.J.J, Aparicio-Maldonado, C. | | Deposit date: | 2021-09-21 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
7S9W
 
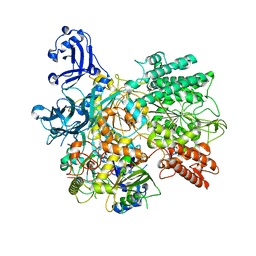 | | Structure of DrmAB:ADP:DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), DrmA, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Brounds, S.J.J, Aparicio-Maldonado, C. | | Deposit date: | 2021-09-21 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
6RRP
 
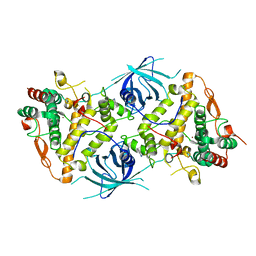 | | Crystal structure of tyrosinase PvdP from Pseudomonas aeruginosa bound to copper and phenylthiourea | | Descriptor: | COPPER (II) ION, N-PHENYLTHIOUREA, PvdP | | Authors: | Wibowo, J.P, Batista, F.A, van Oosterwijk, N, Groves, M.R, Dekker, F.J, Quax, W.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mechanism of inhibition by phenylthiourea on PvdP, a tyrosinase synthesizing pyoverdine of Pseudomonas aeruginosa.
Int.J.Biol.Macromol., 146, 2020
|
|
8HZ5
 
 | |
8HW5
 
 | |
7YQH
 
 | | Cryo-EM structure of 8-subunit Smc5/6 | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 3, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2022-08-07 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
6RZ8
 
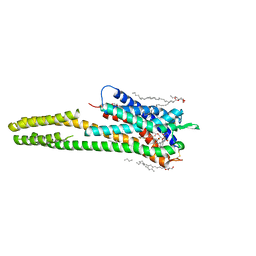 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
7ZSZ
 
 | |
8A9I
 
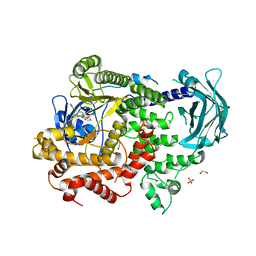 | | PI3KC2a core in complex with PITCOIN1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-~{N}-(1,3-thiazol-2-yl)ethanamide, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
7SIA
 
 | |
7S71
 
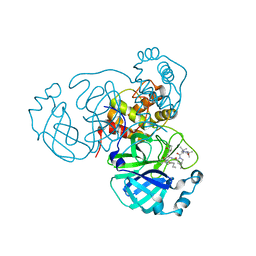 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI35 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S72
 
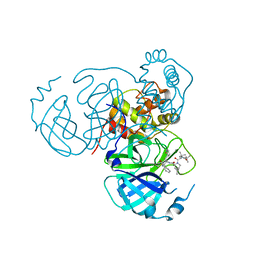 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI36 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6W
 
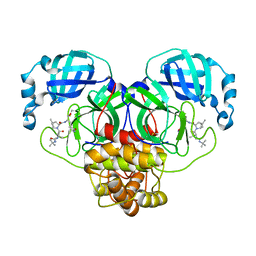 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6X
 
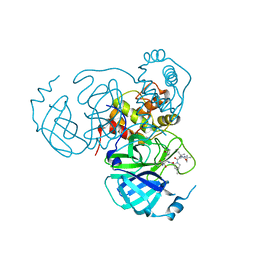 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI30 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
