1C66
 
 | |
5Z26
 
 | |
5Z1Y
 
 | |
5Z2O
 
 | |
5YYC
 
 | | Crystal structure of alanine racemase from Bacillus pseudofirmus (OF4) | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Dong, H, Hu, T.T, He, G.Z, Lu, D.R, Qi, J.X, Dou, Y.S, Long, W, He, X, Su, D, Ju, J.S. | | 登録日 | 2017-12-08 | | 公開日 | 2019-01-02 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural features and kinetic characterization of alanine racemase from Bacillus pseudofirmus OF4.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5ZOD
 
 | | Crystal Structure of hFen1 in apo form | | 分子名称: | Flap endonuclease 1, MAGNESIUM ION, POTASSIUM ION | | 著者 | Han, W, Hua, Y, Zhao, Y. | | 登録日 | 2018-04-13 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOL
 
 | | Crystal structure of a three sites mutantion of FSAA complexed with HA and product | | 分子名称: | (3S,4S)-3,4-dihydroxy-4-(thiophen-2-yl)butan-2-one, 1-hydroxypropan-2-one, CHLORIDE ION, ... | | 著者 | Wu, L, Yang, X.H, Yu, H.W, Zhou, J.H. | | 登録日 | 2018-04-13 | | 公開日 | 2019-06-12 | | 最終更新日 | 2020-07-22 | | 実験手法 | X-RAY DIFFRACTION (2.172 Å) | | 主引用文献 | The engineering of decameric d-fructose-6-phosphate aldolase A by combinatorial modulation of inter- and intra-subunit interactions.
Chem.Commun.(Camb.), 56, 2020
|
|
5ZKY
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 without its lid | | 分子名称: | DFA-IIIase | | 著者 | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | 登録日 | 2018-03-26 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKS
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 | | 分子名称: | DFA-IIIase | | 著者 | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | 登録日 | 2018-03-26 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZOG
 
 | | Crystal Structure of R192F hFen1 in complex with DNA | | 分子名称: | DNA (5'-D(*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), ... | | 著者 | Han, W, Hua, Y, Zhao, Y. | | 登録日 | 2018-04-13 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZMO
 
 | | Sulfur binding domain of ScoMcrA complexed with phosphorothioated DNA | | 分子名称: | DNA (5'-D(P*CP*CP*GP*(GS)P*CP*CP*GP*G)-3'), PHOSPHATE ION, Uncharacterized protein McrA | | 著者 | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | 登録日 | 2018-04-04 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
1TLP
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | 分子名称: | CALCIUM ION, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, THERMOLYSIN, ... | | 著者 | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | 登録日 | 1987-06-29 | | 公開日 | 1989-01-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
6A1I
 
 | | Crystal structure of a synthase 1 from Santalum album | | 分子名称: | CARBONATE ION, GLYCEROL, NICKEL (II) ION, ... | | 著者 | Han, X, Ko, T.P, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-06-07 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a synthase 1 from Santalum album in complex with ligand
To Be Published
|
|
6A2D
 
 | | Crystal structure of a synthase 2 from santalum album in complex with ligand1 | | 分子名称: | MAGNESIUM ION, Sesquisabinene B synthase 2, [bis(chloranyl)-[oxidanyl-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienoxy]phosphoryl]methyl]phosphonic acid | | 著者 | Han, X, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2018-06-10 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal structure of a synthase 2 from santalum album
To be published
|
|
5ZZ6
 
 | | Redox-sensing transcriptional repressor Rex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Redox-sensing transcriptional repressor Rex 1 | | 著者 | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | 登録日 | 2018-05-30 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
6VJQ
 
 | |
5S1A
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-43406 | | 分子名称: | 5-amino-3-methyl-1H-pyrazole-4-carbonitrile, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.079 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
8EWV
 
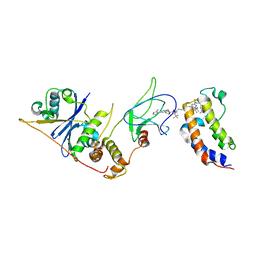 | | DNA-encoded library (DEL)-enabled discovery of proximity inducing small molecules | | 分子名称: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | 著者 | Schreiber, S.L, Shu, W, Ma, X, Michaud, G, Bonazzi, S, Berst, F. | | 登録日 | 2022-10-24 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | DNA-encoded library-enabled discovery of proximity-inducing small molecules.
Nat.Chem.Biol., 20, 2024
|
|
5ZWV
 
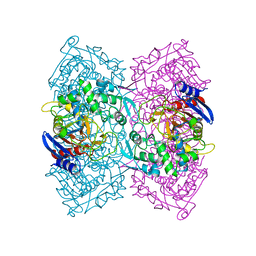 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | 分子名称: | Est-Y29 | | 著者 | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W. | | 登録日 | 2018-05-17 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
6A6P
 
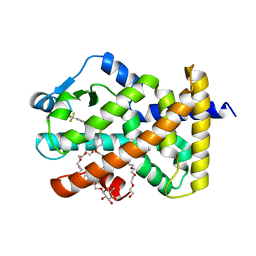 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd)LBD in Complex with DN003316 | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside, ... | | 著者 | Chin, J.W, Cho, S.J, Song, J.Y, Ha, J.H. | | 登録日 | 2018-06-29 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd)LBD in Complex with DN003316
To Be Published
|
|
6A9D
 
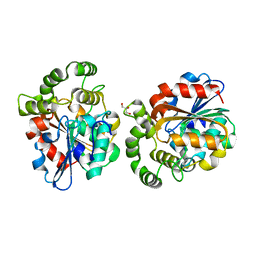 | |
6A9N
 
 | |
5ZVV
 
 | | Structure of SeMet-phAimR | | 分子名称: | AimR transcriptional regulator, GLYCEROL | | 著者 | Cheng, W, Dou, C. | | 登録日 | 2018-05-13 | | 公開日 | 2018-09-05 | | 最終更新日 | 2019-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
6A0W
 
 | | Crystal structure of lipase from Rhizopus microsporus var. chinensis | | 分子名称: | Lipase, SULFATE ION | | 著者 | Zhang, M, Yu, X.W, Xu, Y, Huang, C.H, Guo, R.T. | | 登録日 | 2018-06-06 | | 公開日 | 2019-10-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis by Which the N-Terminal Polypeptide Segment ofRhizopus chinensisLipase Regulates Its Substrate Binding Affinity.
Biochemistry, 58, 2019
|
|
1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | 分子名称: | CALCIUM ION, XYLANASE A | | 著者 | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | 登録日 | 1995-08-31 | | 公開日 | 1996-06-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
