2N9C
 
 | | NRAS Isoform 5 | | 分子名称: | GTPase NRas | | 著者 | Markowitz, J, Mal, T.K, Yuan, C, Courtney, N.B, Patel, M, Stiff, A.R, Blachly, J, Walker, C, Eisfeld, A, de la Chapelle, A, Carson III, W.E. | | 登録日 | 2015-11-13 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of NRAS isoform 5.
Protein Sci., 25, 2016
|
|
8DUS
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (1'-(4-(2-(ethylamino)ethoxy)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | 分子名称: | Estrogen receptor, [(1'R)-1'-{4-[2-(ethylamino)ethoxy]phenyl}-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | 著者 | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | 登録日 | 2022-07-27 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
4RC7
 
 | |
5YPL
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EP complex | | 分子名称: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | 著者 | Feng, H, Wang, D, Liu, W. | | 登録日 | 2017-11-02 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
8DV7
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (6'-hydroxy-1'-(4-(2-(1-propylpyrrolidin-3-yl)ethyl)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | 分子名称: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{2-[(3R)-1-propylpyrrolidin-3-yl]ethyl}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | 著者 | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
6XD0
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-434 | | 分子名称: | 2-[({2-[(2,6-dichlorophenyl)amino]phenyl}acetyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | 登録日 | 2020-06-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.012 Å) | | 主引用文献 | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
8DUG
 
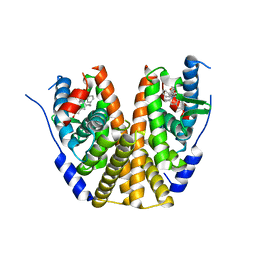 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (6'-hydroxy-1'-(4-(2-((S)-3-methylpyrrolidin-1-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | 分子名称: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{2-[(3R)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | 著者 | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | 登録日 | 2022-07-27 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
8DUB
 
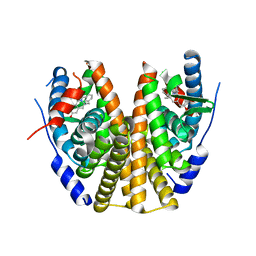 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with ((1'-(4-((1-ethylpyrrolidin-3-yl)methyl)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | 分子名称: | Estrogen receptor, [(1'R)-1'-(4-{[(3S)-1-ethylpyrrolidin-3-yl]oxy}phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | 著者 | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | 登録日 | 2022-07-27 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
6G1T
 
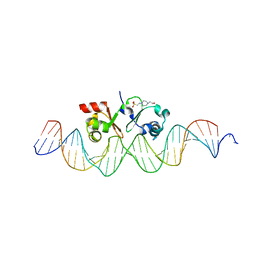 | | TraN, a repressor of an Enterococcus conjugative type IV secretion system | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AM32, DNA (34-MER) | | 著者 | Goessweiner-Mohr, N, Kohler, V, Keller, W. | | 登録日 | 2018-03-22 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | TraN: A novel repressor of an Enterococcus conjugative type IV secretion system.
Nucleic Acids Res., 46, 2018
|
|
6XQT
 
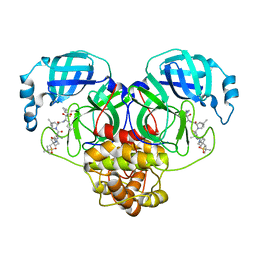 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Narlaprevir | | 分子名称: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kneller, D.W, Kovalevsky, A, Coates, L. | | 登録日 | 2020-07-10 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
6G3E
 
 | | Crystal structure of EDDS lyase in complex with formate | | 分子名称: | Argininosuccinate lyase, FORMIC ACID, SODIUM ION | | 著者 | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | 登録日 | 2018-03-25 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
3PKI
 
 | | Human SIRT6 crystal structure in complex with ADP ribose | | 分子名称: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | 著者 | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-11 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
5XIX
 
 | |
4R57
 
 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, Spermidine n1-acetyltransferase, ... | | 著者 | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-08-20 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.079 Å) | | 主引用文献 | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
7OTA
 
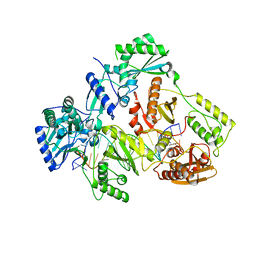 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-230 | | 分子名称: | ((2-(6-amino-9H-purin-9-yl)ethyl)-L-seryl)phosphoramidic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
1U1D
 
 | | Structure of e. coli uridine phosphorylase complexed to 5-(phenylthio)acyclouridine (ptau) | | 分子名称: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLTHIO)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Bu, W, Settembre, E.C, Ealick, S.E. | | 登録日 | 2004-07-15 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7OTN
 
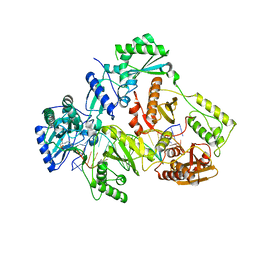 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-247 | | 分子名称: | (S)-2-((2-(6-amino-9H-purin-9-yl)ethyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
8I1O
 
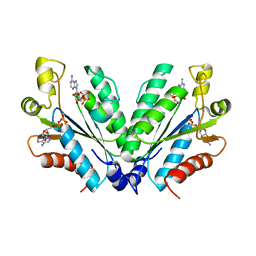 | |
8I1M
 
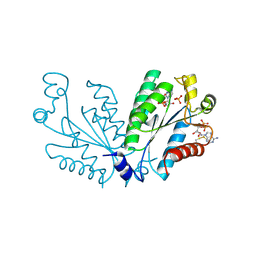 | | Crystal structure of oxidated APSK1 domain from human PAPSS1 in complex with APS and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, PAPSS1 protein, ... | | 著者 | Zhang, L, Song, W.Y, Zhang, L. | | 登録日 | 2023-01-13 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-07-19 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Redox switching mechanism of the adenosine 5'-phosphosulfate kinase domain (APSK2) of human PAPS synthase 2.
Structure, 31, 2023
|
|
8E6D
 
 | | Crystal structure of MERS 3CL protease in complex with a p-fluorophenyl dimethyl sulfane inhibitor | | 分子名称: | (1R,2S)-2-{[N-({2-[(4-fluorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8I1N
 
 | |
7OTZ
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-259 | | 分子名称: | (S)-2-(2-(6-amino-9H-purin-9-yl)ethoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
8E61
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-chlorophenyl dimethyl sulfane inhibitor | | 分子名称: | (1R,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6B
 
 | | Crystal structure of MERS 3CL protease in complex with a dimethyl sulfinyl benzene inhibitor | | 分子名称: | (2~{S})-2-[[(2~{S})-4-methyl-2-[[2-methyl-2-[oxidanyl(phenyl)-$l^{3}-sulfanyl]propoxy]carbonylamino]pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid, N~2~-(ethoxycarbonyl)-N-{(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide, Orf1a protein | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
1TRO
 
 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | 分子名称: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | 著者 | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | 登録日 | 1992-08-30 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
