5GM9
 
 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | 分子名称: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2016-07-13 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
2E76
 
 | | Crystal Structure of the Cytochrome b6f Complex with tridecyl-stigmatellin (TDS) from M.laminosus | | 分子名称: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | 著者 | Cramer, W.A, Yamashita, E, Zhang, H. | | 登録日 | 2007-01-05 | | 公開日 | 2007-06-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
8W4O
 
 | | Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | 分子名称: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | 著者 | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
3HF8
 
 | | Crystal structure of human tryoptophan hydroxylase type 1 with bound LP-533401 and Fe | | 分子名称: | 4-{2-amino-6-[(1R)-2,2,2-trifluoro-1-(3'-fluorobiphenyl-4-yl)ethoxy]pyrimidin-4-yl}-L-phenylalanine, FE (III) ION, Tryptophan 5-hydroxylase 1 | | 著者 | Tari, L.W, Swanson, R.V, Hunter, M.J. | | 登録日 | 2009-05-11 | | 公開日 | 2010-04-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Mechanism of Inhibition of Novel Tryptophan Hydroxylase Inhibitors Revealed by Co-crystal Structures and Kinetic Analysis.
Curr Chem Genomics, 4, 2010
|
|
3DMV
 
 | |
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | 登録日 | 2014-07-23 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.594 Å) | | 主引用文献 | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
3DN2
 
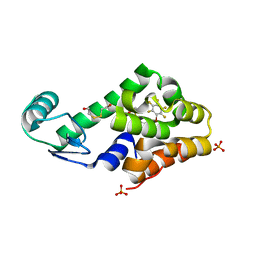 | | Bromopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | 分子名称: | 1-bromo-2,3,4,5,6-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | 著者 | Liu, L, Matthews, B.W. | | 登録日 | 2008-07-01 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
5GJ9
 
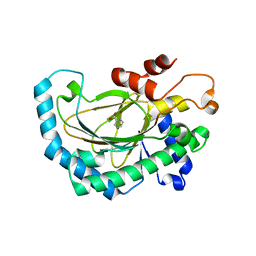 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with POA | | 分子名称: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRAZINE-2-CARBOXYLIC ACID, ZINC ION | | 著者 | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | 登録日 | 2016-06-28 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
3DN8
 
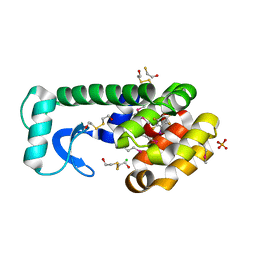 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant (seleno version) | | 分子名称: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, ... | | 著者 | Liu, L, Matthews, B.W. | | 登録日 | 2008-07-01 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
1Y6E
 
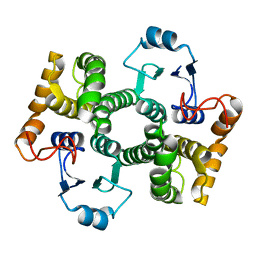 | | Orthorhombic glutathione S-transferase of Schistosoma japonicum | | 分子名称: | glutathione S-transferase | | 著者 | Rufer, A.C, Thiebach, L, Baer, K, Klein, H.W, Hennig, M. | | 登録日 | 2004-12-06 | | 公開日 | 2005-03-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-ray structure of glutathione S-transferase from Schistosoma japonicum in a new crystal form reveals flexibility of the substrate-binding site
Acta Crystallogr.,Sect.F, 61, 2005
|
|
5WGD
 
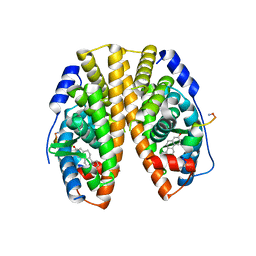 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Estradiol and SRC2-LP1 | | 分子名称: | (ACE)AILHKLLQDS(NH2), (ACE)HKILHKLLQDS(NH2), ESTRADIOL, ... | | 著者 | Fanning, S.W, Speltz, T.E, Mayne, C.G, Siddiqui, Z, Greene, G.L, Tajkhorshid, E, Moore, T.W. | | 登録日 | 2017-07-14 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A "cross-stitched" peptide with improved helicity and proteolytic stability.
Org. Biomol. Chem., 16, 2018
|
|
6WPT
 
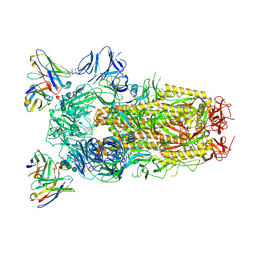 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | 著者 | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
5GTO
 
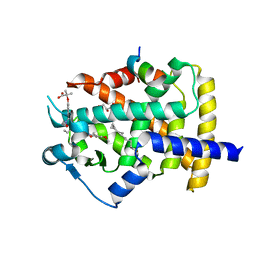 | | Human PPARgamma ligand binding dmain complexed with S35 | | 分子名称: | 2-[4-[5-[(1~{S})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, MYRISTIC ACID, ... | | 著者 | Jang, J.Y, Suh, S.W. | | 登録日 | 2016-08-22 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
3HDZ
 
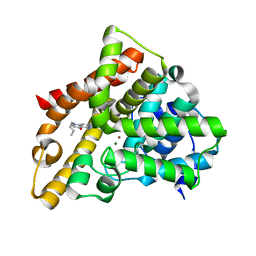 | | Identification, Synthesis, and SAR of Amino Substituted Pyrido[3,2b]pryaziones as Potent and Selective PDE5 Inhibitors | | 分子名称: | 5-amino-1-butyl-7-phenyl-1,6-naphthyridin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | 著者 | Cubbage, J.W, Brown, D.G, Jacobsen, E.J, Walker, J.K, Hughes, R.O. | | 登録日 | 2009-05-07 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification, synthesis and SAR of amino substituted pyrido[3,2b]pyrazinones as potent and selective PDE5 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
3HJB
 
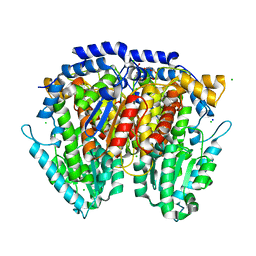 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | 分子名称: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-05-21 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
4RSU
 
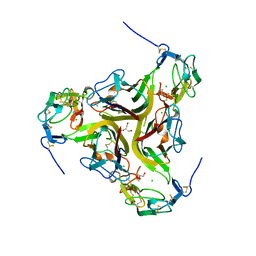 | | Crystal structure of the light and hvem complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, W, Ramagoal, U.A, Himmel, D, Bonanno, J.B, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-11-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
5ZOB
 
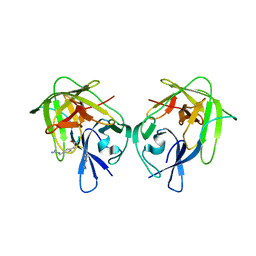 | |
8KC7
 
 | | Rpd3S histone deacetylase complex | | 分子名称: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-06 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
5DRZ
 
 | | Crystal structure of anti-HIV-1 antibody F240 Fab in complex with gp41 peptide | | 分子名称: | Envelope glycoprotein gp160, HIV Antibody F240 Heavy Chain, HIV Antibody F240 Light Chain, ... | | 著者 | Gohain, N, Tolbert, W.D, Pazgier, M. | | 登録日 | 2015-09-16 | | 公開日 | 2016-10-19 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Sci Rep, 6, 2016
|
|
5YYS
 
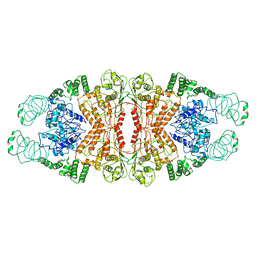 | | Cryo-EM structure of L-fucokinase, GDP-fucose pyrophosphorylase (FKP)in Bacteroides fragilis | | 分子名称: | L-fucokinase, L-fucose-1-P guanylyltransferase | | 著者 | Wang, J, Hu, H, Liu, Y, Zhou, Q, Wu, P, Yan, N, Wang, H.W, Wu, J.W, Sun, L. | | 登録日 | 2017-12-11 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structure of L-fucokinase/GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis.
Protein Cell, 10, 2019
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
