4DQ2
 
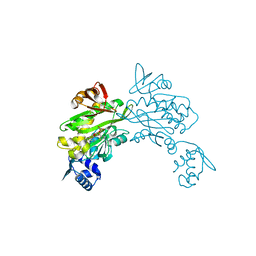 | | Structure of staphylococcus aureus biotin protein ligase in complex with biotinol-5'-amp | | 分子名称: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Biotin-[acetyl-CoA-carboxylase] ligase | | 著者 | Wilce, M, Yap, M, Pendini, N, Soares de Costa, T, Polyak, S, Tieu, W, Booker, G, Wallace, J. | | 登録日 | 2012-02-14 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
7Q5V
 
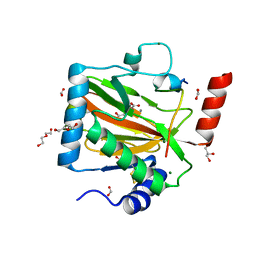 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH N-OXALYLGLYCINE (NOG) AND HIF-2 ALPHA CODD (523-542) | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | 著者 | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | 登録日 | 2021-11-04 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
1CC6
 
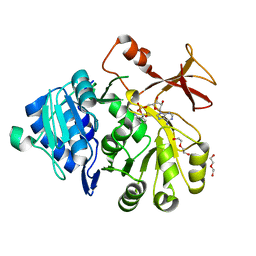
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | 著者 | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | 登録日 | 1999-03-04 | | 公開日 | 1999-03-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
5OE5
 
 | |
5JGJ
 
 | | Crystal structure of GtmA | | 分子名称: | UbiE/COQ5 family methyltransferase, putative | | 著者 | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | 登録日 | 2016-04-20 | | 公開日 | 2017-03-01 | | 最終更新日 | 2018-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
4ZWL
 
 | | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | 分子名称: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-19 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289
To be Published
|
|
4EGM
 
 | | The X-ray crystal structure of CYP199A4 in complex with 4-ethylbenzoic acid | | 分子名称: | 4-ethylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | 登録日 | 2012-03-31 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
6FKH
 
 | | Chloroplast F1Fo conformation 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | 著者 | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | 登録日 | 2018-01-24 | | 公開日 | 2018-05-23 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
1PAU
 
 | |
4ZT5
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (2S)-N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)-2-methylpropane-1,3-diamine (Chem 1655) | | 分子名称: | (2S)-N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)-2-methylpropane-1,3-diamine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Koh, C.-Y, Hol, W.G.J. | | 登録日 | 2015-05-14 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | 5-Fluoroimidazo[4,5-b]pyridine Is a Privileged Fragment That Conveys Bioavailability to Potent Trypanosomal Methionyl-tRNA Synthetase Inhibitors.
Acs Infect Dis., 2, 2016
|
|
5IJ8
 
 | | Structure of the primary oncogenic mutant Y641N Hs/AcPRC2 in complex with a pyridone inhibitor | | 分子名称: | 5,8-dichloro-2-[(4-ethyl-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-({1-[(2R)-2-hydroxypropanoyl]piperidin-4-yl}oxy)-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of Zeste Homolog 2 (EZH2),Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | 著者 | Gajiwala, K.S, Brooun, A, Deng, Y.-L, Liu, W. | | 登録日 | 2016-03-01 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.
Nat Commun, 7, 2016
|
|
1KS3
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | 登録日 | 2002-01-10 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
7U69
 
 | |
7U3M
 
 | |
2WTI
 
 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | 分子名称: | 1,2-ETHANEDIOL, 4-[2-AMINO-5-(2,3-DIHYDROTHIENO[3,4-B][1,4]DIOXIN-5-YL)PYRIDIN-3-YL]BENZAMIDE, CHECKPOINT KINASE 2, ... | | 著者 | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | 登録日 | 2009-09-16 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
7U6A
 
 | |
4EHG
 
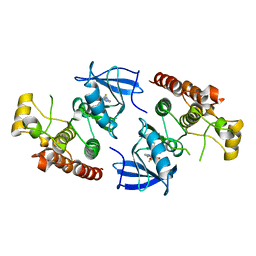 | | B-Raf Kinase Domain in Complex with an Aminopyridimine-based Inhibitor | | 分子名称: | N-{2,4-difluoro-3-[({6-[(2-hydroxyethyl)amino]pyrimidin-4-yl}carbamoyl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | 著者 | Voegtli, W.C. | | 登録日 | 2012-04-02 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Potent and selective aminopyrimidine-based B-raf inhibitors with favorable physicochemical and pharmacokinetic properties.
J.Med.Chem., 55, 2012
|
|
7U6B
 
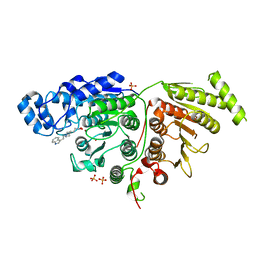 | |
8W4P
 
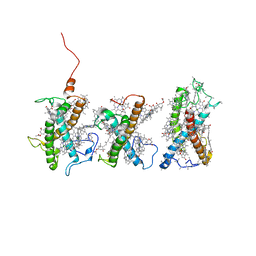 | | Structure of PSII-FCPII-I/J/K complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | 分子名称: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, ... | | 著者 | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
3ODM
 
 | | Archaeal-type phosphoenolpyruvate carboxylase | | 分子名称: | GOLD (I) CYANIDE ION, MALONATE ION, Phosphoenolpyruvate carboxylase | | 著者 | Dunten, P.W. | | 登録日 | 2010-08-11 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure of an archaeal-type phosphoenolpyruvate carboxylase sensitive to inhibition by aspartate.
Proteins, 79, 2011
|
|
4R4Q
 
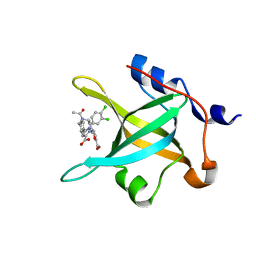 | | Crystal structure of RPA70N in complex with 5-(3-((N-(4-(5-carboxyfuran-2-yl)benzyl)acetamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | 分子名称: | 5-[4-({acetyl[4-(5-carboxyfuran-2-yl)benzyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | 著者 | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | 登録日 | 2014-08-19 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
8W4O
 
 | | Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | 分子名称: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | 著者 | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
5O1P
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase. | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | 著者 | Kopec, J, Rembeza, E, Pena, I.A, Williams, E, Velupillai, S, Kupinska, K, Strain-Damerell, C, Goubin, S, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | 登録日 | 2017-05-18 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase.
To Be Published
|
|
3ZVI
 
 | | Methylaspartate ammonia lyase from Clostridium tetanomorphum mutant L384A | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Raj, H, Szymanski, W, de Villiers, J, Rozeboom, H.J, Veetil, V.P, Reis, C.R, de Villiers, M, de Wildeman, S, Dekker, F.J, Quax, W.J, Thunnissen, A.M.W.H, Feringa, B.L, Janssen, D.B, Poelarends, G.J. | | 登録日 | 2011-07-25 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Nat.Chem., 4, 2012
|
|
1PPO
 
 | | DETERMINATION OF THE STRUCTURE OF PAPAYA PROTEASE OMEGA | | 分子名称: | MERCURY (II) ION, PROTEASE OMEGA | | 著者 | Pickersgill, R.W, Rizkallah, P.J, Harris, G.W, Goodenough, P.W. | | 登録日 | 1991-07-12 | | 公開日 | 1993-10-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Determination of the Structure of Papaya Protease Omega
Acta Crystallogr.,Sect.B, 47, 1991
|
|
