2PPS
 
 | | PHOTOSYNTHETIC REACTION CENTER AND CORE ANTENNA SYSTEM (TRIMERIC), ALPHA CARBON ONLY | | 分子名称: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHOTOSYSTEM I, ... | | 著者 | Krauss, N, Schubert, W.-D, Klukas, O, Fromme, P, Witt, H.T, Saenger, W. | | 登録日 | 1997-05-27 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Photosystem I at 4 A resolution represents the first structural model of a joint photosynthetic reaction centre and core antenna system.
Nat.Struct.Biol., 3, 1996
|
|
6H1L
 
 | | Structure of the BM3 heme domain in complex with tioconazole | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | 著者 | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | 登録日 | 2018-07-12 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.968 Å) | | 主引用文献 | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
4Z8N
 
 | |
262L
 
 | |
2FEQ
 
 | | orally active thrombin inhibitors | | 分子名称: | Decapeptide Hirudin Analogue, N-(CARBOXYMETHYL)-3-CYCLOHEXYL-D-ALANYL-N-({4-[(E)-AMINO(IMINO)METHYL]-1,3-THIAZOL-2-YL}METHYL)-L-PROLINAMIDE, Thrombin heavy chain, ... | | 著者 | Mack, H, Baucke, D, Hornberger, W, Lange, U.E.W, Hoeffken, H.W. | | 登録日 | 2005-12-16 | | 公開日 | 2006-08-08 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Orally active thrombin inhibitors. Part 1: optimization of the P1-moiety
Bioorg.Med.Chem.Lett., 16, 2006
|
|
201L
 
 | |
229L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GUANIDINE, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-26 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
3CPA
 
 | |
2OVO
 
 | |
8XZF
 
 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | 分子名称: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZJ
 
 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | 分子名称: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
5KU6
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and methazolamide | | 分子名称: | ACETATE ION, Carbonic anhydrase 4, GLYCEROL, ... | | 著者 | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | 登録日 | 2016-07-12 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
8XZH
 
 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | 分子名称: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
1WA1
 
 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | 著者 | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | 登録日 | 2004-10-22 | | 公開日 | 2005-01-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
8XZI
 
 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | 分子名称: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
7ZX0
 
 | | Crystal structure of Pol theta polymerase domain in complex with compound 5 | | 分子名称: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-[5-bromanyl-3-cyano-6-methyl-4-(trifluoromethyl)pyridin-2-yl]oxy-~{N}-ethyl-~{N}-(3-methylphenyl)ethanamide, DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*(DDG))-3'), ... | | 著者 | Krajewski, W.W, Turnbull, A.P, Willis, S, Charles, M, Stockley, M, Heald, R.A. | | 登録日 | 2022-05-19 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Discovery, Characterization, and Structure-Based Optimization of Small-Molecule In Vitro and In Vivo Probes for Human DNA Polymerase Theta.
J.Med.Chem., 65, 2022
|
|
8WSW
 
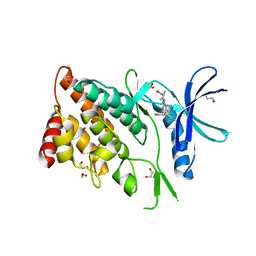 | | The Crystal Structure of LIMK2a from Biortus | | 分子名称: | 1,2-ETHANEDIOL, LIM domain kinase 2, ~{N}-[5-[2-[2,6-bis(chloranyl)phenyl]-5-[bis(fluoranyl)methyl]pyrazol-3-yl]-1,3-thiazol-2-yl]-2-methyl-propanamide | | 著者 | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | 登録日 | 2023-10-17 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Crystal Structure of LIMK2a from Biortus.
To Be Published
|
|
7ZUS
 
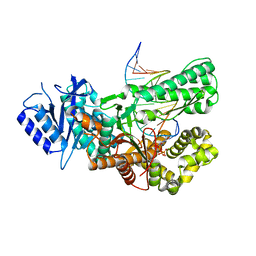 | | Crystal structure of ternary complex of Pol theta polymerase domain | | 分子名称: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*(DDG))-3'), DNA (5'-D(P*TP*TP*CP*CP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*C)-3'), ... | | 著者 | Krajewski, W.W, Turnbull, A.P, Willis, S, Charles, M, Stockley, M, Heald, R.A. | | 登録日 | 2022-05-13 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Discovery, Characterization, and Structure-Based Optimization of Small-Molecule In Vitro and In Vivo Probes for Human DNA Polymerase Theta.
J.Med.Chem., 65, 2022
|
|
2SCU
 
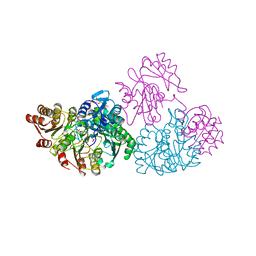 | | A detailed description of the structure of Succinyl-COA synthetase from Escherichia coli | | 分子名称: | COENZYME A, PROTEIN (SUCCINYL-COA LIGASE), SULFATE ION | | 著者 | Fraser, M.E, Wolodko, W.T, James, M.N.G, Bridger, W.A. | | 登録日 | 1998-09-24 | | 公開日 | 1999-08-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A detailed structural description of Escherichia coli succinyl-CoA synthetase.
J.Mol.Biol., 285, 1999
|
|
6XOG
 
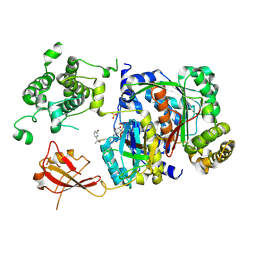 | | Structure of SUMO1-ML786519 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6LIG
 
 | |
6XOH
 
 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.226 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
5IMN
 
 | | Crystal structure of N299A/S303A Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | 分子名称: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | 著者 | Chen, M, Christianson, D.W. | | 登録日 | 2016-03-06 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.527 Å) | | 主引用文献 | Probing the Role of Active Site Water in the Sesquiterpene Cyclization Reaction Catalyzed by Aristolochene Synthase.
Biochemistry, 55, 2016
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6LRS
 
 | | Cryo-EM structure of RbcL8-RbcS4 from Anabaena sp. PCC 7120 | | 分子名称: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | 著者 | Xia, L.Y, Jiang, Y.L, Kong, W.W, Sun, H, Li, W.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2020-01-16 | | 公開日 | 2020-07-15 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
