2BHX
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure A) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIG
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure I) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
4B0M
 
 | | Complex of the Caf1AN usher domain, Caf1M chaperone and Caf1 subunit from Yersinia pestis | | Descriptor: | CHAPERONE PROTEIN CAF1M, F1 CAPSULE ANTIGEN, F1 CAPSULE-ANCHORING PROTEIN | | Authors: | Dubnovitsky, A, Yu, X.D, Pudney, A.F, MacIntyre, S, Knight, S.D, Zavialov, A.V. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric Mechanism Controls Traffic in the Chaperone/Usher Pathway.
Structure, 20, 2012
|
|
2BI3
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure D) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2VRY
 
 | | Mouse Neuroglobin with heme iron in the reduced ferrous state | | Descriptor: | NEUROGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Arcovito, A, Moschetti, T, D'Angelo, P, Mancini, G, Vallone, B, Brunori, M, Della Longa, S. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An X-Ray Diffraction and X-Ray Absorption Spectroscopy Joint Study of Neuroglobin.
Arch.Biochem.Biophys., 475, 2008
|
|
2JHO
 
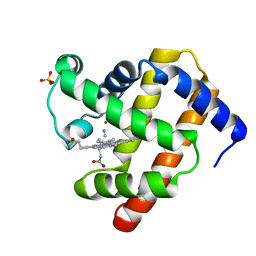 | | Cyanomet Sperm Whale Myoglobin at 1.4A resolution | | Descriptor: | CYANIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Arcovito, A, Benfatto, M, Cianci, M, Hasnain, S.S, Nienhaus, K, Nienhaus, G.U, Savino, C, Strange, R.W, Vallone, B, Della Longa, S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure analysis of a metalloprotein with enhanced active-site resolution using in situ x-ray absorption near edge structure spectroscopy.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
8BCJ
 
 | |
8BCI
 
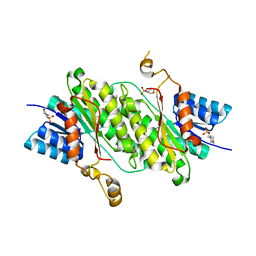 | |
8ZBO
 
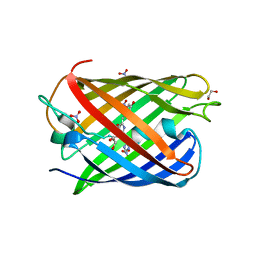 | | Crystal structure of the biphotochromic fluorescent protein moxSAASoti (F97M variant) in its green on-state | | Descriptor: | 1,2-ETHANEDIOL, F97M variant of the biphotochromic fluorescent protein moxSAASoti, NITRATE ION, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Marynich, N.K, Minyaev, M.E, Khadiyatova, A.A, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-point substitution F97M leads to in cellulo crystallization of the biphotochromic protein moxSAASoti.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
6O2P
 
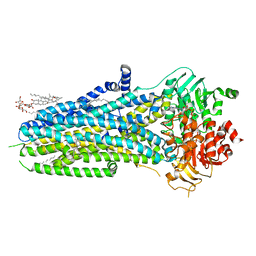 | | Complex of ivacaftor with cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Liu, F, Zhang, Z, Chen, J, Levit, A, Shoichet, B. | | Deposit date: | 2019-02-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of a hotspot on CFTR for potentiation.
Science, 364, 2019
|
|
8C18
 
 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
6O1V
 
 | | Complex of human cystic fibrosis transmembrane conductance regulator (CFTR) and GLPG1837 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Zhang, Z, Liu, F, Chen, J, Levit, A, Shoichet, B. | | Deposit date: | 2019-02-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural identification of a hotspot on CFTR for potentiation.
Science, 364, 2019
|
|
8PEI
 
 | | Crystal structure of the biphotochromic fluorescent protein SAASoti (C21N/V127T variant) in its green on-state | | Descriptor: | C21N/V127T form of the biphotochromic fluorescent protein SAASoti | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Matyuta, I.O, Gavshina, A.V, Solovyev, I.D, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of the correlated motion(s) of the chromophore in photoswitching of green and red forms of the photoconvertible fluorescent protein mSAASoti.
Sci Rep, 14, 2024
|
|
8QHS
 
 | | Cryo-EM structure of the monocin tail-tube, MttP. | | Descriptor: | Antigen A | | Authors: | Nadejda, S, Lichtenstein, R, Schlussel, S, Azulay, G, Borovok, I, Holdengraber, V, Elad, N, Wolf, S.G, Zalk, R, Zarivach, R, Frank, G.A, Herskovits, A.A. | | Deposit date: | 2023-09-10 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Tailocin cell factories
To Be Published
|
|
8SMG
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMD
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, ABC transporter ATPase | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMF
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Gp34.65, MAGNESIUM ION | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SME
 
 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8V9M
 
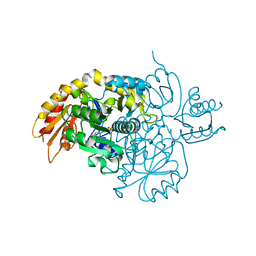 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (R)-3-amino-5,5-difluorocyclohex-1-ene-1-carboxylic acid. | | Descriptor: | 3-fluoro-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]benzoic acid, GLYCEROL, Ornithine aminotransferase, ... | | Authors: | Vargas, A.L, Devitt, A, Kaley, N, Silverman, R, Liu, D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, and Mechanistic Studies of ( R )-3-Amino-5,5-difluorocyclohex-1-ene-1-carboxylic Acid as an Inactivator of Human Ornithine Aminotransferase.
Acs Chem.Biol., 19, 2024
|
|
5NI9
 
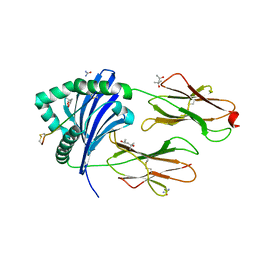 | | Crystal structure of HLA-DRB1*04:01 with the alpha-enolase peptide 326-340 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-enolase, ... | | Authors: | Gerstner, C, Dubnovitsky, A. | | Deposit date: | 2017-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Memory T cells specific to citrullinated alpha-enolase are enriched in the rheumatic joint.
J. Autoimmun., 92, 2018
|
|
5NIG
 
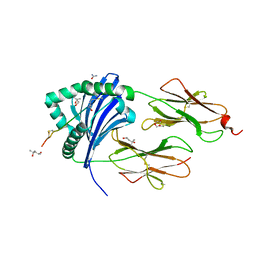 | |
7UAV
 
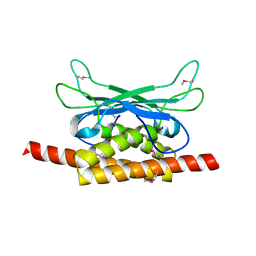 | | Structure of Clostridium botulinum prophage Tad1 in apo state | | Descriptor: | ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
7UAW
 
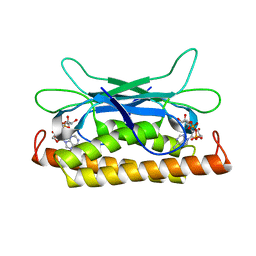 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
5TVN
 
 | | Crystal structure of the LSD-bound 5-HT2B receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, S, McCorvy, J.D, Betz, R.M, Venkatakrishnan, A.J, Levit, A, Lansu, K, Schools, Z.L, Che, T, Nichols, D.E, Shoichet, B.K, Dror, R.O, Roth, B.L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of an LSD-Bound Human Serotonin Receptor.
Cell, 168, 2017
|
|
6CM4
 
 | | Structure of the D2 Dopamine Receptor Bound to the Atypical Antipsychotic Drug Risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, D(2) dopamine receptor, endolysin chimera, ... | | Authors: | Wang, S, Che, T, Levit, A, Shoichet, B.K, Wacker, D, Roth, B.L. | | Deposit date: | 2018-03-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone.
Nature, 555, 2018
|
|
