4B9X
 
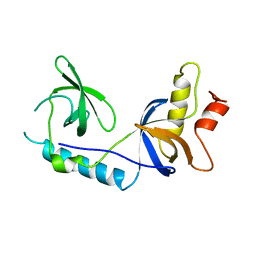 | | Structure of extended Tudor domain TD3 from mouse TDRD1 | | 分子名称: | TUDOR DOMAIN-CONTAINING PROTEIN 1 | | 著者 | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | 登録日 | 2012-09-08 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Multiple Tudor Domain-Containing Protein Tdrd1 is a Molecular Scaffold for Mouse Piwi Proteins and Pirna Biogenesis Factors.
RNA, 18, 2012
|
|
8FZ9
 
 | |
4C1Q
 
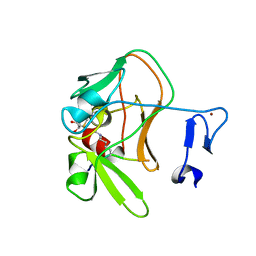 | | Crystal structure of the PRDM9 SET domain in complex with H3K4me2 and AdoHcy. | | 分子名称: | GLYCEROL, HISTONE H3.1, HISTONE-LYSINE N-METHYLTRANSFERASE PRDM9, ... | | 著者 | Mathioudakis, N, Cusack, S, Kadlec, J. | | 登録日 | 2013-08-13 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular Basis for the Regulation of the H3K4 Methyltransferase Activity of Prdm9.
Cell Rep., 5, 2013
|
|
6H77
 
 | | E1 enzyme for ubiquitin like protein activation in complex with UBL | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | 登録日 | 2018-07-30 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
6H78
 
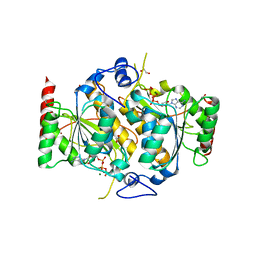 | | E1 enzyme for ubiquitin like protein activation. | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | 登録日 | 2018-07-30 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
8TCI
 
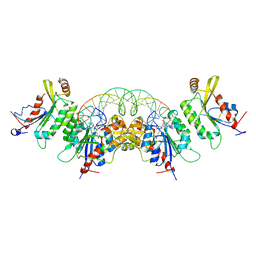 | | Crystal structure of DNMT3C-DNMT3L in complex with CGG DNA | | 分子名称: | (5'-D(P*CP*AP*TP*G)-R(P*(PYO))-D(P*GP*GP*TP*CP*TP*AP*AP*TP*TP*AP*GP*AP*CP*CP*GP*CP*AP*TP*G)-3'), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3C, ... | | 著者 | Khudaverdyan, N, Song, J. | | 登録日 | 2023-07-01 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | The structure of DNA methyltransferase DNMT3C reveals an activity-tuning mechanism for DNA methylation.
J.Biol.Chem., 2024
|
|
4B9W
 
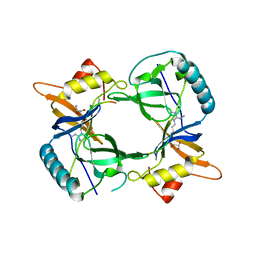 | | Structure of extended Tudor domain TD3 from mouse TDRD1 in complex with MILI peptide containing dimethylarginine 45. | | 分子名称: | GLYCEROL, PIWI-LIKE PROTEIN 2, TUDOR DOMAIN-CONTAINING PROTEIN 1 | | 著者 | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | 登録日 | 2012-09-08 | | 公開日 | 2012-10-17 | | 最終更新日 | 2019-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors.
Rna, 18, 2012
|
|
3X43
 
 | | Crystal structure of O-ureido-L-serine synthase | | 分子名称: | O-ureido-L-serine synthase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | 登録日 | 2015-03-13 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
3X44
 
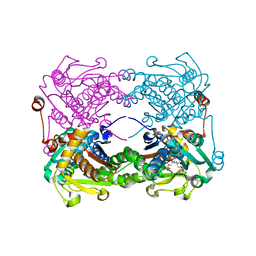 | | Crystal structure of O-ureido-L-serine-bound K43A mutant of O-ureido-L-serine synthase | | 分子名称: | (E)-O-(carbamoylamino)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, O-ureido-L-serine synthase | | 著者 | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | 登録日 | 2015-03-13 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
6DBY
 
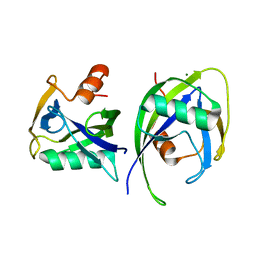 | | Crystal structure of Nudix 1 from Arabidopsis thaliana | | 分子名称: | MAGNESIUM ION, Nudix hydrolase 1 | | 著者 | Noel, J.P, Thomas, S.T, Dudareva, N, Henry, L.K. | | 登録日 | 2018-05-03 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Contribution of isopentenyl phosphate to plant terpenoid metabolism.
Nat Plants, 4, 2018
|
|
6DBZ
 
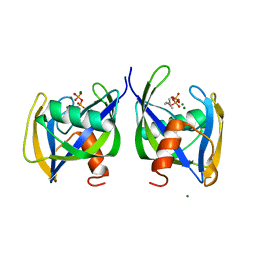 | | Crystal structure of Nudix 1 from Arabidopsis thaliana complexed with isopentenyl diphosphate | | 分子名称: | ISOPENTYL PYROPHOSPHATE, MAGNESIUM ION, Nudix hydrolase 1 | | 著者 | Noel, J.P, Thomas, S.T, Dudareva, N, Henry, L.K. | | 登録日 | 2018-05-03 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Contribution of isopentenyl phosphate to plant terpenoid metabolism.
Nat Plants, 4, 2018
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | 分子名称: | BolA-like protein RIKEN cDNA 1110025L05 | | 著者 | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-01-26 | | 公開日 | 2004-02-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
5CU5
 
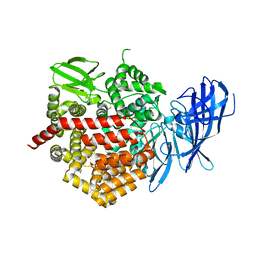 | | Crystal structure of ERAP2 without catalytic Zn(II) atom | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | 著者 | Saridakis, E, Mathioudakis, N, Giastas, P, Mavridis, I.M, Stratikos, E. | | 登録日 | 2015-07-24 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
6FUC
 
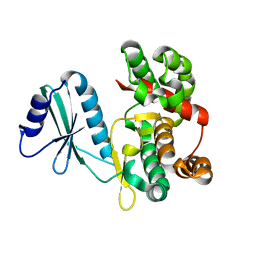 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 | | 分子名称: | Aminoglycoside phosphotransferase | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | 登録日 | 2018-02-26 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6FUX
 
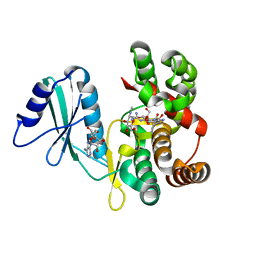 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 in complex with ADP and streptomycin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, GLYCEROL, ... | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | 登録日 | 2018-02-28 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
8HYF
 
 | | Crystal Structure of Banana Lectin In-complex with Fucose at 2.95 A Resolution | | 分子名称: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Rasheed, S, Arif, R, Huda, N, Ahmad, M.S, Mateen, S.M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal Structure of Banana Lectin in-Complex with L-Fucose at 2.95 A Resolution
To Be Published
|
|
8IGF
 
 | | Crystal Structure of Human Carbonic Anhydrase II In-complex with 4-Acetylphenylboronic acid at 2.6 A Resolution | | 分子名称: | (4-ethanoylphenyl)boronic acid, Carbonic anhydrase 2, GLYCEROL, ... | | 著者 | Rasheed, S, Huda, N, Fisher, S.Z, Falke, S, Gul, S, Ahmad, M.S, Choudhary, M.I. | | 登録日 | 2023-02-20 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification, crystallization, and first X-ray structure analyses of phenyl boronic acid-based inhibitors of human carbonic anhydrase-II.
Int.J.Biol.Macromol., 267, 2024
|
|
8J2O
 
 | |
8JEE
 
 | | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution | | 分子名称: | Carbonic anhydrase 2, GLYCEROL, Levosulpiride, ... | | 著者 | Rasheed, S, Huda, N, Falke, S, Fisher, S.Z, Ahmad, M.S. | | 登録日 | 2023-05-15 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution
To Be Published
|
|
5Y5N
 
 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | 分子名称: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | 著者 | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | 登録日 | 2017-08-09 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
1SYK
 
 | | Crystal structure of E230Q mutant of cAMP-dependent protein kinase reveals unexpected apoenzyme conformation | | 分子名称: | cAMP-dependent protein kinase, alpha-catalytic subunit | | 著者 | Wu, J, Yang, J, Madhusudan, N, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the E230Q mutant of cAMP-dependent
protein kinase reveals an unexpected apoenzyme conformation and an
extended N-terminal A helix.
Protein Sci., 14, 2005
|
|
