1XN9
 
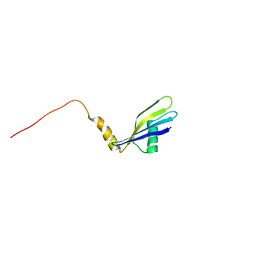 | | Solution Structure of Methanosarcina mazei Protein RPS24E: The Northeast Structural Genomics Consortium Target MaR11 | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Liu, G, Xiao, R, Parish, D, Ma, L, Sukumaran, D, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methanosarcina mazei Protein RPS24E: The Northeast Structural Genomics Consortium Target MaR11
To be Published
|
|
1XPV
 
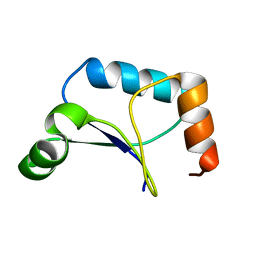 | | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris | | Descriptor: | hypothetical protein XCC2852 | | Authors: | Shao, Y, Acton, T.B, Liu, G, Ma, L, Shen, Y, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris
To be Published
|
|
1XM3
 
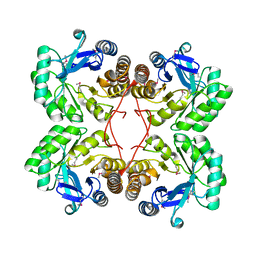 | | Crystal structure of Northeast Structural Genomics Target SR156 | | Descriptor: | Thiazole biosynthesis protein thiG | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S, Forouhar, F, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Northeast Structural Genomics Target SR156
To be Published
|
|
4NZG
 
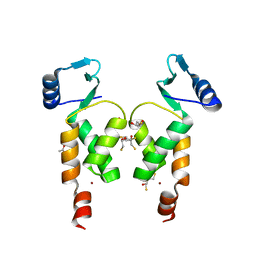 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Integrase p46, ... | | Authors: | Guan, R, Jiang, M, Janjua, H, Maglaqui, M, Zhao, L, Xiao, R, Acton, T.B, Everett, J.K, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
1NY1
 
 | | CRYSTAL STRUCTURE OF B. SUBTILIS POLYSACCHARIDE DEACETYLASE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR127. | | Descriptor: | Probable polysaccharide deacetylase pdaA | | Authors: | Forouhar, F, Edstrom, W, Khan, J, Ma, L, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Polysaccharide Deacetylase (PDAA_BACSU) from B. Subtilis (Pdaa_Bacsu) Northeast Structural Genomics Research Consortium (Nesg) Target Sr127
To be Published
|
|
4N0B
 
 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1XHJ
 
 | | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630. Northest Structural Genomics Consortium Target SeR8. | | Descriptor: | Nitrogen Fixation Protein NifU | | Authors: | Baran, M.C, Huang, Y.P, Acton, T, Xiao, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630.
Northest Strucutral Genomics Consortium Target SeR8.
To be Published
|
|
1NXZ
 
 | | X-Ray Crystal Structure of Protein yggj_haein of Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR73. | | Descriptor: | Hypothetical protein HI0303 | | Authors: | Forouhar, F, Shen, J, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional assignment based on structural analysis: crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot.
Proteins, 53, 2003
|
|
1NXU
 
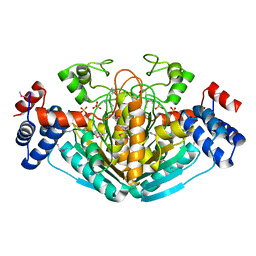 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
1XQB
 
 | | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC)target IR47. | | Descriptor: | Hypothetical UPF0066 protein HI0510 | | Authors: | Benach, J, Lee, I, Forouhar, F, Kuzin, A.P, Keller, J.P, Itkin, A, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC) target IR47.
To be Published
|
|
2KW2
 
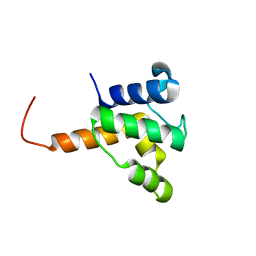 | | Solution NMR of the specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Rossi, P, Lee, H, Wohlbold, T.J, Valafar, H, Lemak, A, Ertekin, A, Forouhar, F, Ciccosanti, C, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-30 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
1YUD
 
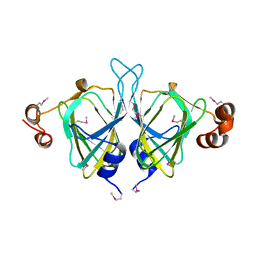 | | X-ray Crystal Structure of Protein SO0799 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR12. | | Descriptor: | hypothetical protein SO0799 | | Authors: | Kuzin, A.P, Vorobiev, S, Chen, Y, Forouhar, F, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein SO0799 from Shewanella oneidensis
To be Published
|
|
4WVC
 
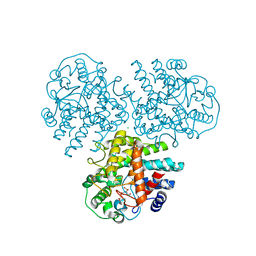 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4WVA
 
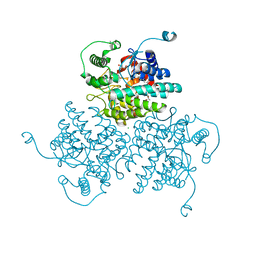 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
2L6U
 
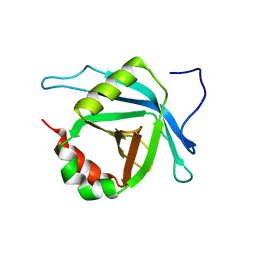 | | Solution NMR Structure of Med25(391-543) Comprising the Activator-Interacting Domain (ACID) of Human Mediator Subuniti 25. Northeast Structural Genomics Consortium Target HR6188A | | Descriptor: | Mediator complex subunit MED25 | | Authors: | Eletsky, A, Ryuechan, W.T, Sukumaran, D.K, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of MED25(391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25.
J.Struct.Funct.Genom., 12, 2011
|
|
4WVB
 
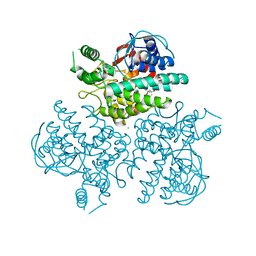 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Uncharacterized protein, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
8EAZ
 
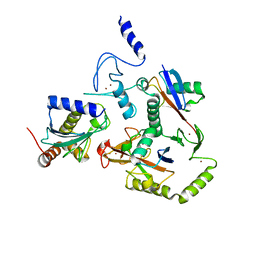 | | HOIL-1/E2-Ub/Ub transthiolation complex | | Descriptor: | RanBP-type and C3HC4-type zinc finger-containing protein 1, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Wang, X.S, Cotton, T.R, Lechtenberg, B.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family.
Nat Commun, 14, 2023
|
|
2ICT
 
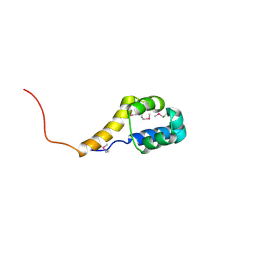 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
4XPO
 
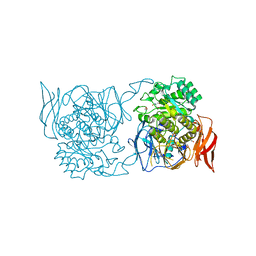 | | Crystal structure of a novel alpha-galactosidase from Pedobacter saltans | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-glucosidase | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPR
 
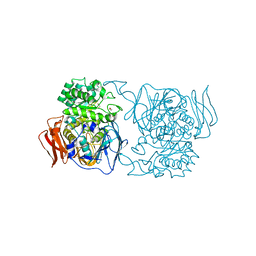 | | Crystal structure of the mutant D365A of Pedobacter saltans GH31 alpha-galactosidase | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
2M9W
 
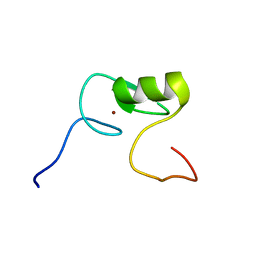 | | Solution NMR Structure of Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B | | Descriptor: | Transcription factor GATA-4, ZINC ION | | Authors: | Xu, X, Eletsky, A, Lee, D, Kohn, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B
To be Published
|
|
4GPM
 
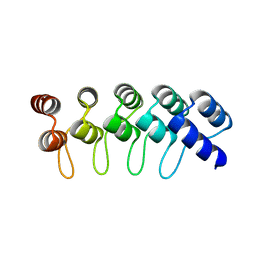 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR264. | | Descriptor: | Engineered Protein OR264 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
4XPP
 
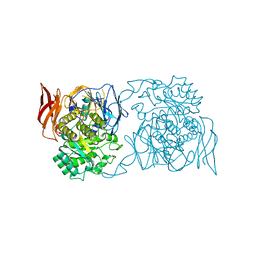 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with D-galactose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase, beta-D-galactopyranose | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPQ
 
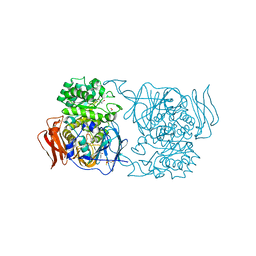 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with L-fucose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-glucosidase, ... | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
