4LGD
 
 | | Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ras association domain family member 5, ... | | Authors: | Luo, X, Ni, L, Tomchick, D.R. | | Deposit date: | 2013-06-27 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5.
Structure, 21, 2013
|
|
4LG4
 
 | |
5TVF
 
 | | Crystal structure of Trypanosoma brucei AdoMetDC/prozyme heterodimer in complex with inhibitor CGP 40215 | | Descriptor: | 1,4-DIAMINOBUTANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[C-[N'-(3-CARBAMIMIDOYL-BENZYLIDENIUM)-HYDRAZINO]-[[AMINOMETHYLIDENE]AMINIUM]-IMINOMETHYL]-BENZAMIDINIUM, ... | | Authors: | Phillips, M.A, Volkov, O.A, Chen, Z, Tomchick, D.R. | | Deposit date: | 2016-11-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
3LET
 
 | | Crystal Structure of Fic domain containing AMPylator, VopS | | Descriptor: | Adenosine monophosphate-protein transferase vopS | | Authors: | Luong, P.H, Kinch, L.N, Brautigam, C.A, Grishin, N.V, Tomchick, D.R, Orth, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural and Kinetic Analysis of VopS with Fic Domain Supports a Direct Transfer Mechanism for AMPylation
To be Published
|
|
5TVO
 
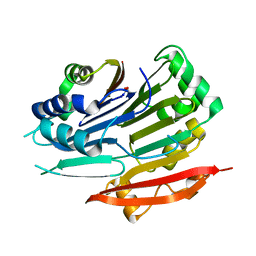 | | Crystal structure of Trypanosoma brucei AdoMetDC-delta26 monomer | | Descriptor: | S-adenosylmethionine decarboxylase alpha chain, S-adenosylmethionine decarboxylase proenzyme, SODIUM ION | | Authors: | Volkov, O.A, Ariagno, C, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
5U2P
 
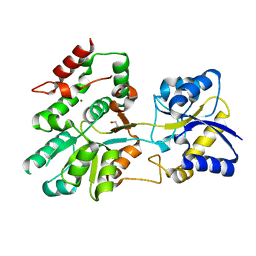 | | The crystal structure of Tp0737 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Norgard, M.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Functional clues from the crystal structure of an orphan periplasmic ligand-binding protein from Treponema pallidum.
Protein Sci., 26, 2017
|
|
8USO
 
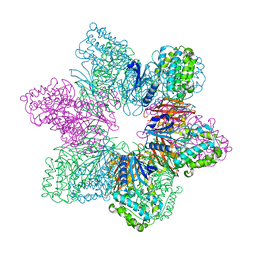 | | Full-length human CaMKII delta holoenzyme | | Descriptor: | MALONATE ION, calcium/calmodulin-dependent protein kinase | | Authors: | Ozden, C, Abromson, N.L, Tomchick, D.R, Stratton, M.M, Garman, S.C. | | Deposit date: | 2023-10-28 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human CaMKII delta holoenzyme
To be published
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
4GGD
 
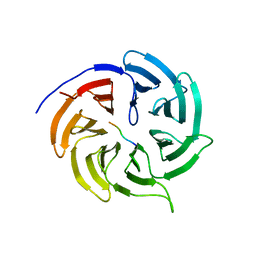 | |
4GGC
 
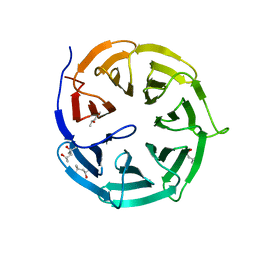 | |
4GGA
 
 | |
2OIV
 
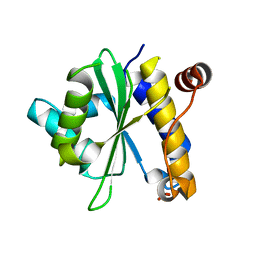 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
2O6E
 
 | | Structure of native rTp34 from Treponema pallidum from zinc-soaked crystals | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2O6F
 
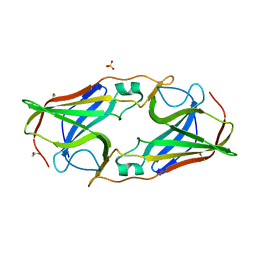 | | Structure of metal- free rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2O6D
 
 | | Structure of native rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2O6C
 
 | | Structure of selenomethionyl rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2OIX
 
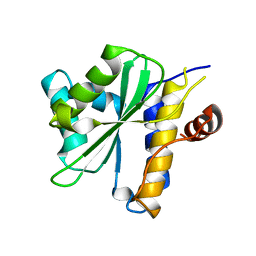 | | Xanthomonas XopD C470A Mutant | | Descriptor: | Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
3EDV
 
 | |
1U3D
 
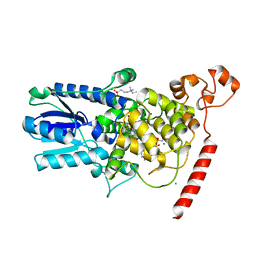 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3F1O
 
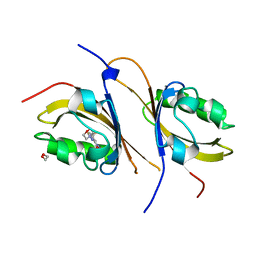 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with an internally-bound artificial ligand | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, ... | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F1N
 
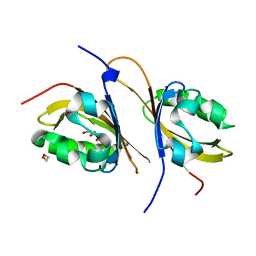 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with internally bound ethylene glycol. | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F1P
 
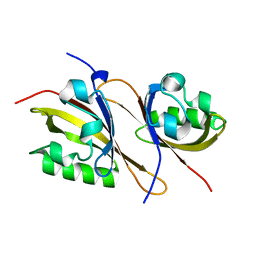 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1U3C
 
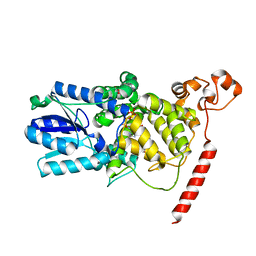 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6MRU
 
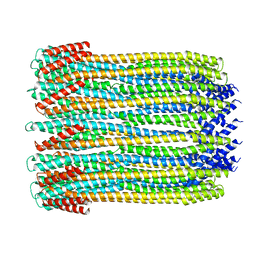 | | 13-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
