1UHI
 
 | | Crystal structure of i-aequorin | | Descriptor: | (2R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(4-IODOBENZYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHH
 
 | | Crystal structure of cp-aequorin | | Descriptor: | (8R)-8-(CYCLOPENTYLMETHYL)-2-HYDROPEROXY-2-(4-HYDROXYBENZYL)-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H) -ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHJ
 
 | | Crystal structure of br-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-(4-BROMOBENZYL)-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHK
 
 | | Crystal structure of n-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(2-NAPHTHYLMETHYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
4RSP
 
 | |
4YLU
 
 | | X-ray structure of MERS-CoV nsp5 protease bound with a non-covalent inhibitor | | Descriptor: | ACETATE ION, N-{4-[(1H-benzotriazol-1-ylacetyl)(thiophen-3-ylmethyl)amino]phenyl}propanamide, ORF1a protein | | Authors: | Tomar, S, Mesecar, A.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): IMPLICATIONS FOR nsp5 REGULATION AND THE DEVELOPMENT OF ANTIVIRALS.
J.Biol.Chem., 290, 2015
|
|
1Z0Q
 
 | | Aqueous Solution Structure of the Alzheimer's Disease Abeta Peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Tomaselli, S, Esposito, V, Vangone, P, van Nuland, N.A, Bonvin, A.M, Guerrini, R, Tancredi, T, Temussi, P.A, Picone, D. | | Deposit date: | 2005-03-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The alpha-to-beta Conformational Transition of Alzheimer's Abeta-(1-42) Peptide in Aqueous Media is Reversible: A Step by Step Conformational Analysis Suggests the Location of beta Conformation Seeding
Chembiochem, 7, 2006
|
|
3H7I
 
 | | Structure of the metal-free D132N T4 RNase H | | Descriptor: | Ribonuclease H, SULFATE ION | | Authors: | Tomanicek, S.J, Mueser, T.C. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Additional Order Appears in the Absence of Metals in a FEN-1 protein: Structural Analysis of Magnesium Binding to Bacteriophage T4 RNaseH
To be Published
|
|
3H8J
 
 | |
2LFO
 
 | | NMR structure of cl-BABP/SS complexed with glycochenodeoxycholic and glycocholic acids | | Descriptor: | Fatty acid-binding protein, liver, GLYCOCHENODEOXYCHOLIC ACID, ... | | Authors: | Tomaselli, S, Cogliati, C, Pagano, K, Zetta, L, Zanzoni, S, Assfalg, M, Molinari, H, Ragona, L. | | Deposit date: | 2011-07-07 | | Release date: | 2012-07-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A disulfide bridge allows for site-selective binding in liver bile acid binding protein thereby stabilising the orientation of key amino acid side chains.
Chemistry, 18, 2012
|
|
2K62
 
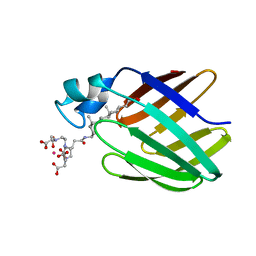 | | NMR solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based Gd(III)-chelate | | Descriptor: | (3alpha,5alpha,8alpha)-3-[(N,N-bis{2-[bis(carboxymethyl)amino]ethyl}-L-gamma-glutamyl)amino]cholan-24-oic acid, Liver fatty acid-binding protein, YTTERBIUM (III) ION | | Authors: | Tomaselli, S, Zanzoni, S, Ragona, L, Gianolio, E, Aime, S, Assfalg, M, Molinari, H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based gadolinium(III)-chelate, a potential hepatospecific magnetic resonance imaging contrast agent.
J.Med.Chem., 51, 2008
|
|
2XR0
 
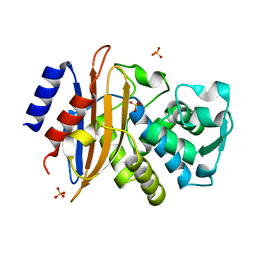 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
3H8S
 
 | |
3H8W
 
 | |
2X7V
 
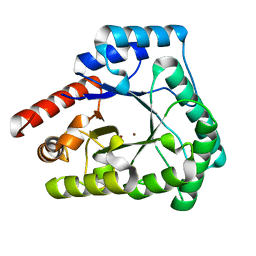 | | Crystal structure of Thermotoga maritima endonuclease IV in the presence of zinc | | Descriptor: | PROBABLE ENDONUCLEASE 4, ZINC ION | | Authors: | Tomanicek, S.J, Hughes, R.C, Ng, J.D, Coates, L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Endonuclease Iv Homologue from Thermotoga Maritima in the Presence of Active-Site Divalent Metal Ions
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2WYX
 
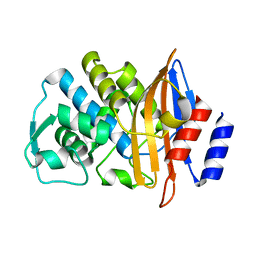 | | Neutron structure of a class A Beta-lactamase Toho-1 E166A R274N R276N triple mutant | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Blakeley, M.P, Cooper, J, Chen, Y, Afonine, P, Coates, L. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Neutron Diffraction Studies of a Class a Beta-Lactamase Toho-1 E166A R274N R276N Triple Mutant
J.Mol.Biol., 396, 2010
|
|
2X7W
 
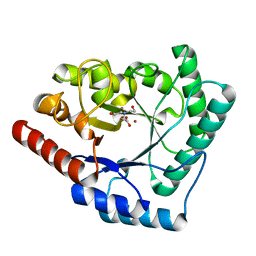 | | Crystal structure of Thermotoga maritima endonuclease IV in the presence of cadmium and zinc | | Descriptor: | BICINE, CADMIUM ION, PROBABLE ENDONUCLEASE 4, ... | | Authors: | Tomanicek, S.J, Hughes, R.C, Ng, J.D, Coates, L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the Endonuclease Iv Homologue from Thermotoga Maritima in the Presence of Active-Site Divalent Metal Ions
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2XQZ
 
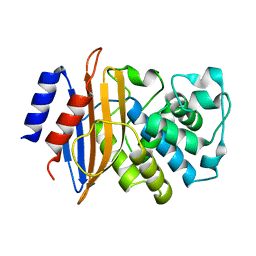 | | Neutron structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
4BD0
 
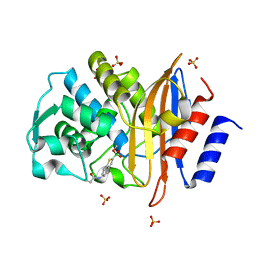 | | X-ray structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
2GVS
 
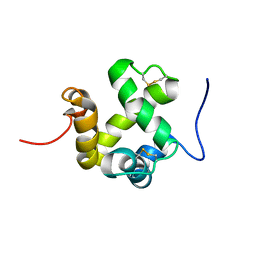 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
4BD1
 
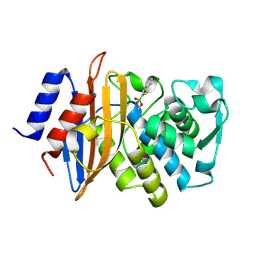 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-16 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.002 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
1WSV
 
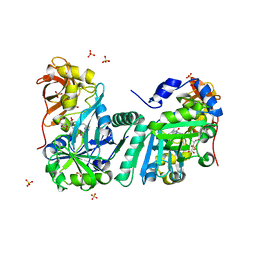 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-11 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1WSR
 
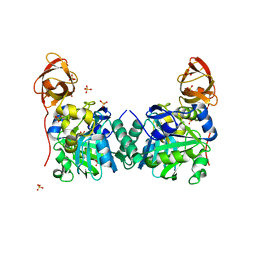 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1X2H
 
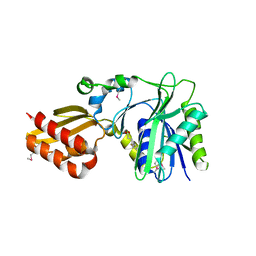 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
6JY0
 
 | | CryoEM structure of S.typhimurium R-type straight flagellar filament made of FljB (A461V) | | Descriptor: | Flagellin | | Authors: | Yamaguchi, T, Toma, S, Terahara, N, Miyata, T, Minamino, T, Ashikara, M, Namba, K, Kato, T. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural and Functional Comparison ofSalmonellaFlagellar Filaments Composed of FljB and FliC.
Biomolecules, 10, 2020
|
|
