1MSV
 
 | | The S68A S-adenosylmethionine decarboxylase proenzyme processing mutant. | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Tolbert, W.D, Zhang, Y, Bennett, E.M, Cottet, S.E, Ekstrom, J.L, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of Human S-Adenosylmethionine
Decarboxylase Proenzyme Processing as Revealed by the
Structure of the S68A Mutant.
Biochemistry, 42, 2003
|
|
5KJR
 
 | |
6MFP
 
 | | Crystal Structure of the RV305 C1-C2 specific ADCC potent antibody DH677.3 Fab in complex with HIV-1 clade A/E gp120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Young, B, Pazgier, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.
J.Virol., 94, 2020
|
|
1N13
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AGMATINE, Pyruvoyl-dependent arginine decarboxylase alpha chain, ... | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-16 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1N2M
 
 | | The S53A Proenzyme Structure of Methanococcus jannaschii. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Pyruvoyl-dependent arginine decarboxylase | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-23 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1MT1
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannaschii | | Descriptor: | AGMATINE, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE ALPHA CHAIN, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE BETA CHAIN | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
8DCQ
 
 | |
2QJ2
 
 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QJ4
 
 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8SM2
 
 | |
8TTW
 
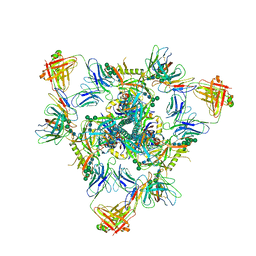 | | Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tolbert, W.D, Pozharski, E, Pazgier, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
4YBL
 
 | |
4YC2
 
 | |
7N0X
 
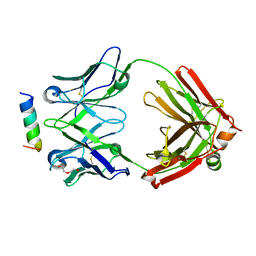 | | Rhesusized RV144 DH827 Fab bound to HIV-1 Env V2 peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycoprotein 120, Rhesusized RV144 DH827 heavy chain Fab fragment, ... | | Authors: | Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-05-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Fc-Effector Function of Rhesusized Variants of Human Anti-HIV-1 IgG1s.
Front Immunol, 12, 2021
|
|
7N8Q
 
 | |
6OZ4
 
 | |
6OZ2
 
 | |
7KCZ
 
 | | CRYSTAL STRUCTURE OF RHESUS MACAQUE (MACACA MULATTA) IGG1 FC FRAGMENT- FC-GAMMA RECEPTOR III COMPLEX V158 MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, ... | | Authors: | Tolbert, W.D, Pazgier, M. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques.
Front Immunol, 13, 2022
|
|
7KJM
 
 | |
7KJN
 
 | |
7S4S
 
 | |
3HN4
 
 | |
3HMR
 
 | |
7SX7
 
 | |
7SX6
 
 | |
