2Q9P
 
 | | Human diphosphoinositol polyphosphate phosphohydrolase 1, Mg-F complex | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, FLUORIDE ION, ... | | Authors: | Thorsell, A.G, Busam, R, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Van den Berg, S, Weigelt, J, Welin, M, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-13 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human diphosphoinositol phosphatase 1.
Proteins, 77, 2009
|
|
4UXB
 
 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, POLY ADP-RIBOSE POLYMERASE 1, SULFATE ION | | Authors: | Tresaugues, L, Thorsell, A.G, Karlberg, T, Schuler, H. | | Deposit date: | 2014-08-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4R6E
 
 | | Human artd1 (parp1) - catalytic domain in complex with inhibitor niraparib | | Descriptor: | 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Karlberg, T, Thorsell, A.G, Brock, J, Schuler, H. | | Deposit date: | 2014-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J.Med.Chem., 60, 2017
|
|
2VUX
 
 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
2VR2
 
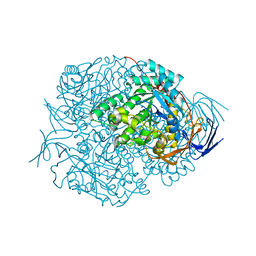 | | Human Dihydropyrimidinase | | Descriptor: | CHLORIDE ION, DIHYDROPYRIMIDINASE, ZINC ION | | Authors: | Welin, M, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Human Dihydropyrimidinase
To be Published
|
|
2IBN
 
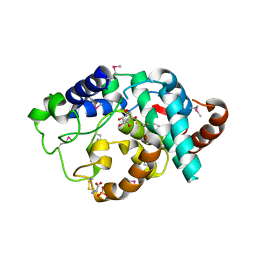 | | Crystal structure of Human myo-Inositol Oxygenase (MIOX) | | Descriptor: | (2S,3R,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXANONE, CYSTEINE, FE (III) ION, ... | | Authors: | Hallberg, B.M, Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Thorsell, A.G, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biophysical Characterization of Human myo-Inositol Oxygenase
J.Biol.Chem., 283, 2008
|
|
4X52
 
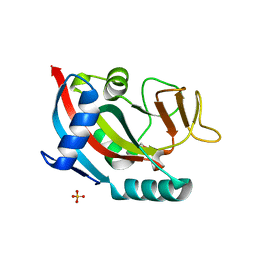 | | Human PARP13 (ZC3HAV1), C-Terminal PARP Domain (H810N; N830Y variant) | | Descriptor: | GLYCEROL, SULFATE ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Karlberg, T, Thorsell, A.G, Klepsch, M, Schuler, H. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
3F0W
 
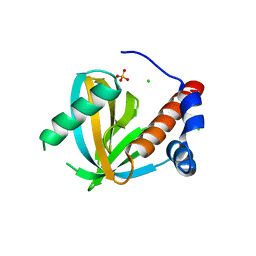 | | Human NUMB-like protein, phosphotyrosine interaction domain | | Descriptor: | CHLORIDE ION, Numb-like protein, SULFATE ION | | Authors: | Lehtio, L, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, D Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human NUMB-like protein, phosphotyrosine interaction domain
To be Published
|
|
3ELB
 
 | | Human CTP: Phosphoethanolamine Cytidylyltransferase in complex with CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Ethanolamine-phosphate cytidylyltransferase, GLYCEROL | | Authors: | Karlberg, T, Welin, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wikstrom, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-22 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human CTP:Phosphoethanolamine Cytidylyltransferase
To be Published
|
|
3EQ5
 
 | | Crystal structure of fragment 137 to 238 of the human Ski-like protein | | Descriptor: | Ski-like protein | | Authors: | Tresaugues, L, Wisniewska, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Van Den Berg, S, Welin, M, Wikstrom, M, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of fragment 137 to 238 of the human Ski-like protein.
To be Published
|
|
3EC8
 
 | | The crystal structure of the RA domain of FLJ10324 (RADIL) | | Descriptor: | CHLORIDE ION, GLYCEROL, LEAD (II) ION, ... | | Authors: | Wisniewska, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the RA domain of FLJ10324 (RADIL)
to be published
|
|
6EJP
 
 | | Yersinia YscU C-terminal fragment in complex with a synthetic compound | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Karlberg, T, Thorsell, A.G, Ho, O, Sunduru, N, Elofsson, M, Wolf-Watz, M, Schuler, H. | | Deposit date: | 2017-09-22 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Yersinia YscU C-terminal fragment in complex with a synthetic compound
To Be Published
|
|
5NQE
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with an N-aryl piperazine inhibitor | | Descriptor: | 3-[[4-[4-(4-fluorophenyl)piperazin-1-yl]-4-oxidanylidene-butanoyl]amino]benzamide, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Design and synthesis of potent inhibitors of the mono(ADP-ribosyl)transferase, PARP14.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4RV6
 
 | |
4TVJ
 
 | | HUMAN ARTD2 (PARP2) - CATALYTIC DOMAIN IN COMPLEX WITH OLAPARIB | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Pinto, A.F, Schuler, H. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4UND
 
 | | HUMAN ARTD1 (PARP1) - CATALYTIC DOMAIN IN COMPLEX WITH INHIBITOR TALAZOPARIB | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, POLY [ADP-RIBOSE] POLYMERASE 1, SODIUM ION | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Klepsch, M, Pinto, A.F, Tresaugues, L, Moche, M, Schuler, H. | | Deposit date: | 2014-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5LX6
 
 | |
4MXE
 
 | | Human ESCO1 (Eco1/Ctf7 ortholog), acetyltransferase domain in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase ESCO1 | | Authors: | Karlberg, T, Wisniewska, M, Thorsell, A.G, Kouznetsova, E, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Svensson, L, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-26 | | Release date: | 2015-04-08 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sister Chromatid Cohesion Establishment Factor ESCO1 Operates by Substrate-Assisted Catalysis.
Structure, 24, 2016
|
|
5LYH
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor H10 | | Descriptor: | 3-[2-[4-[2-[[4-[(3-aminocarbonylphenyl)amino]-4-oxidanylidene-butanoyl]amino]ethyl]-1,2,3-triazol-1-yl]ethylsulfamoyl]benzoic acid, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Small Molecule Microarray Based Discovery of PARP14 Inhibitors.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LXP
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor H5 | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}'-(3-aminocarbonylphenyl)-~{N}-[[1-[(2~{R})-2-phenylpropyl]-1,2,3-triazol-4-yl]methyl]pentanediamide | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2016-09-22 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Small Molecule Microarray Based Discovery of PARP14 Inhibitors.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4R5W
 
 | | Human artd1 (parp1) - catalytic domain in complex with inhibitor xav939 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J.Med.Chem., 60, 2017
|
|
2VXO
 
 | | Human GMP synthetase in complex with XMP | | Descriptor: | GMP SYNTHASE [GLUTAMINE-HYDROLYZING], SULFATE ION, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Welin, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P. | | Deposit date: | 2008-07-08 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity and Oligomerization of Human Gmp Synthetase
J.Mol.Biol., 425, 2013
|
|
2WGH
 
 | | Human Ribonucleotide reductase R1 subunit (RRM1) in complex with dATP and Mg. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Welin, R.M, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh-Nielsen, T, Kotzsch, A, Kotenyova, T, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-04-19 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Allosteric Regulation of Human Ribonucleotide Reductase by Nucleotide-Induced Oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2I4I
 
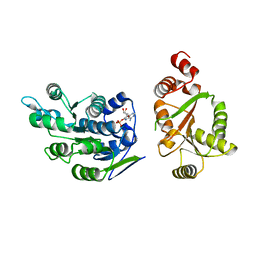 | | Crystal Structure of human DEAD-box RNA helicase DDX3X | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DDX3X | | Authors: | Hogbom, M, Karlberg, T, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP
J.Mol.Biol., 372, 2007
|
|
2J4E
 
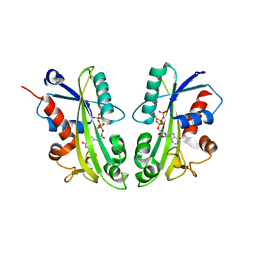 | | THE ITP COMPLEX OF HUMAN INOSINE TRIPHOSPHATASE | | Descriptor: | INOSINE 5'-TRIPHOSPHATE, INOSINE TRIPHOSPHATE PYROPHOSPHATASE, INOSINIC ACID, ... | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmbergschiavone, L, Hogbom, M, Kotenyova, T, Landry, R, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Schuler, H, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Inosine Triphosphatase: Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
