1MIU
 
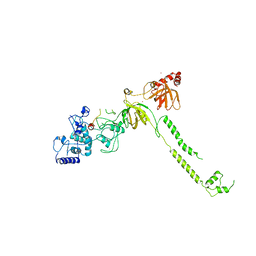 | | Structure of a BRCA2-DSS1 complex | | Descriptor: | Breast Cancer type 2 susceptibility protein, Deleted in split hand/split foot protein 1, MERCURY (II) ION | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure
Science, 297, 2002
|
|
2YB8
 
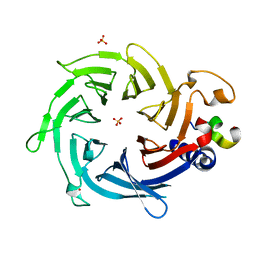 | | Crystal structure of Nurf55 in complex with Su(z)12 | | Descriptor: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
1E1C
 
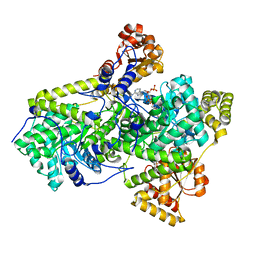 | | METHYLMALONYL-COA MUTASE H244A Mutant | | Descriptor: | COBALAMIN, DESULFO-COENZYME A, METHYLMALONYL-COA MUTASE ALPHA CHAIN, ... | | Authors: | Evans, P.R, Thoma, N.H. | | Deposit date: | 2000-04-30 | | Release date: | 2000-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Protection of Radical Intermediates at the Active Site of Adenosylcobalamin-Dependent Methylmalonyl-Coa Mutase
Biochemistry, 39, 2000
|
|
2YBA
 
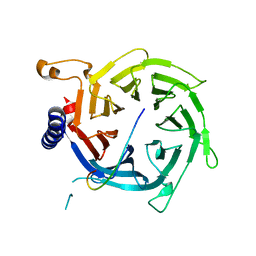 | | Crystal structure of Nurf55 in complex with histone H3 | | Descriptor: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
1MJE
 
 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
2FXO
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FXM
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | MERCURY (II) ION, Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3EI2
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp abasic site containing DNA-duplex | | Descriptor: | 5'-D(*DAP*DAP*DAP*DTP*DGP*DAP*DAP*DTP*(3DR)P*DAP*DAP*DGP*DCP*DAP*DGP*DG)-3', 5'-D(*DCP*DCP*DTP*DGP*DCP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DTP*DT)-3', DNA damage-binding protein 1, ... | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI4
 
 | | Structure of the hsDDB1-hsDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Scrima, A, Pavletich, N.P, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI3
 
 | | Structure of the hsDDB1-drDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI1
 
 | |
6R91
 
 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
 | | Cryo-EM structure of NCP-6-4PP | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6TD3
 
 | | Structure of DDB1 bound to CR8-engaged CDK12-cyclinK | | Descriptor: | (2R)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Bunker, R.D, Petzold, G, Kozicka, Z, Thoma, N.H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K.
Nature, 585, 2020
|
|
6T90
 
 | | OCT4-SOX2-bound nucleosome - SHL-6 | | Descriptor: | DNA (146-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-10-25 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6TBL
 
 | |
6TBN
 
 | | Crystal structure of CIAO1-CIAO2B CIA core complex | | Descriptor: | MIP18 family protein galla-2, Probable cytosolic iron-sulfur protein assembly protein Ciao1, SODIUM ION | | Authors: | Kassube, S.A, Thoma, N.H. | | Deposit date: | 2019-11-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into Fe-S protein biogenesis by the CIA targeting complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TC0
 
 | | Crystal structure of MMS19-CIAO1-CIAO2B CIA targeting complex | | Descriptor: | MIP18 family protein galla-2, MMS19 nucleotide excision repair protein homolog, Probable cytosolic iron-sulfur protein assembly protein Ciao1 | | Authors: | Kassube, S.A, Thoma, N.H. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into Fe-S protein biogenesis by the CIA targeting complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T93
 
 | | Nucleosome with OCT4-SOX2 motif at SHL-6 | | Descriptor: | DNA (153-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-10-25 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6R8Z
 
 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
 | | Cryo-EM structure of NCP_THF2(-3) | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R90
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
8BUR
 
 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
