4IIG
 
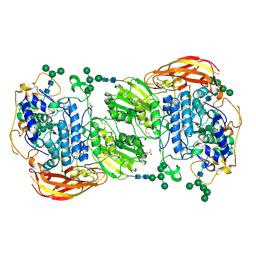 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with D-glucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIC
 
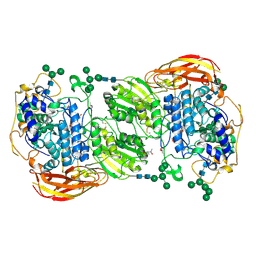 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with isofagomine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIE
 
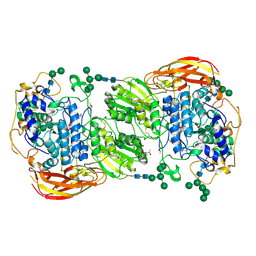 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with calystegine B(2) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIF
 
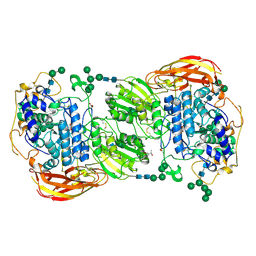 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with castanospermine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
3P8C
 
 | | Structure and Control of the Actin Regulatory WAVE Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Abl interactor 2, CHLORIDE ION, ... | | Authors: | Chen, Z, Borek, D, Padrick, S.B, Gomez, T.S, Metlagel, Z, Ismail, A.M, Umetani, J, Billadeau, D.D, Otwinowski, Z, Rosen, M.K. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure and control of the actin regulatory WAVE complex.
Nature, 468, 2010
|
|
1IEH
 
 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
7E3X
 
 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
7E24
 
 | |
7E28
 
 | |
2HZ0
 
 | | Abl kinase domain in complex with NVP-AEG082 | | Descriptor: | 2-{[(6-OXO-1,6-DIHYDROPYRIDIN-3-YL)METHYL]AMINO}-N-[4-PROPYL-3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2HZ4
 
 | | Abl kinase domain unligated and in complex with tetrahydrostaurosporine | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
1A9M
 
 | | G48H MUTANT OF HIV-1 PROTEASE IN COMPLEX WITH A PEPTIDIC INHIBITOR U-89360E | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Zhang, X.-J, Foundling, S, Hartsuck, J.A, Tang, J. | | Deposit date: | 1998-04-08 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a G48H mutant of HIV-1 protease explains how glycine-48 replacements produce mutants resistant to inhibitor drugs.
FEBS Lett., 420, 1997
|
|
3QPI
 
 | | Crystal Structure of Dimeric Chlorite Dismutases from Nitrobacter winogradskyi | | Descriptor: | Chlorite Dismutase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mlynek, G, Sjoeblom, B, Kostan, J, Fuereder, S, Maixner, F, Furtmueller, P.G, Obinger, O, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2011-02-13 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi.
J.Bacteriol., 193, 2011
|
|
7E8L
 
 | | The structure of Spodoptera litura chemosensory protein | | Descriptor: | Putative chemosensory protein CSP8 | | Authors: | Xie, W, Jia, Q, Zeng, H, Xiao, N, Tang, J, Gao, S, Zhang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Spodoptera litura Chemosensory Protein CSP8.
Insects, 12, 2021
|
|
2RU8
 
 | | DnaT C-terminal domain | | Descriptor: | Primosomal protein 1 | | Authors: | Abe, Y, Tani, J, Fujiyama, S, Urabe, M, Sato, K, Aramaki, T, Katayama, T, Ueda, T. | | Deposit date: | 2014-01-29 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of the primosome protein DnaT-functional structures for homotrimerization, dissociation of ssDNA from the PriB·ssDNA complex, and formation of the DnaT·ssDNA complex.
Febs J., 281, 2014
|
|
2HYY
 
 | | Human Abl kinase domain in complex with imatinib (STI571, Glivec) | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
3OCV
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase mutant D66N complexed with 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
4ACM
 
 | | CDK2 IN COMPLEX WITH 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}-PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE | | Descriptor: | 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, CYCLIN-DEPENDENT KINASE 2, GLYCEROL | | Authors: | Berg, S, Bhat, R, Anderson, M, Bergh, M, Brassington, C, Hellberg, S, Jerning, E, Hogdin, K, Lo-Alfredsson, Y, Neelissen, J, Nilsson, Y, Ormo, M, Soderman, P, Stanway, J, Tucker, J, von Berg, S, Weigelt, T, Xue, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
2G94
 
 | | Crystal structure of beta-secretase bound to a potent and highly selective inhibitor. | | Descriptor: | Beta-secretase 1, N~2~-[(2R,4S,5S)-5-{[N-{[(3,5-DIMETHYL-1H-PYRAZOL-1-YL)METHOXY]CARBONYL}-3-(METHYLSULFONYL)-L-ALANYL]AMINO}-4-HYDROXY-2,7-DIMETHYLOCTANOYL]-N-ISOBUTYL-L-VALINAMIDE | | Authors: | Hong, L, Ghosh, A, Tang, J. | | Deposit date: | 2006-03-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design, synthesis and X-ray structure of protein-ligand complexes: important insight into selectivity of memapsin 2 (beta-secretase) inhibitors.
J.Am.Chem.Soc., 128, 2006
|
|
5G3L
 
 | | ESCHERICHIA COLI HEAT LABILE ENTEROTOXIN TYPE IIB B-PENTAMER COMPLEXED WITH SIALYLATED SUGAR | | Descriptor: | HEAT-LABILE ENTEROTOXIN IIB, B CHAIN, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Zalem, D, Benktander, J, Ribeiro, J.P, Varrot, A, Lebens, M, Imberty, A, Teneberg, S. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Biochemical and Structural Characterization of the Novel Sialic Acid-Binding Site of Escherichia Coli Heat-Labile Enterotoxin Lt-Iib.
Biochem.J., 473, 2016
|
|
4DPD
 
 | | WILD TYPE PLASMODIUM FALCIPARUM DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE (PfDHFR-TS), DHF COMPLEX, NADP+, dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, DIHYDROFOLIC ACID, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4IIH
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with thiocellobiose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
3OCY
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase Complexed with inorganic phosphate | | Descriptor: | Lipoprotein E, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OCU
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase mutant D66N complexed with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
4DP3
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P218 and NADPH | | Descriptor: | 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
