8W1D
 
 | | CRYSTAL STRUCTURE OF DPS-LIKE PROTEIN PA4880 FROM PSEUDOMONAS AERUGINOSA (DIMERIC FORM) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION | | Authors: | Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8FFD
 
 | | Crystal structure of manganeese bound Dps protein (PA0962) from Pseudomonas aeruginosa (cubic form) | | Descriptor: | L(+)-TARTARIC ACID, MANGANESE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FF9
 
 | | Crystal structure of Apo Dps protein (PA0962) from Pseudomonas aeruginosa (orthorhombic form) | | Descriptor: | CHLORIDE ION, Probable dna-binding stress protein, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FFA
 
 | |
8FFC
 
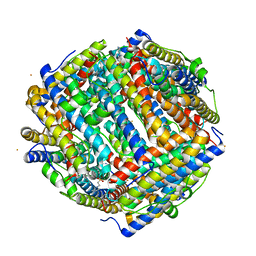 | | Crystal structure of iron bound Dps protein (PA0962) from Pseudomonas aeruginosa (cubic form) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FFB
 
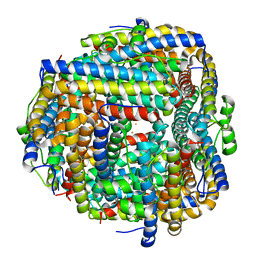 | | Crystal structure of iron bound Dps protein (PA0962) from Pseudomonas aeruginosa (orthorhombic form) | | Descriptor: | FE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FCH
 
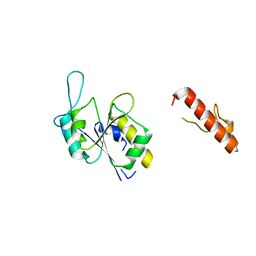 | | Apo Structure of (N1G37) Methyltransferase from Mycobacterium avium | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Isaacson, B, Brylewski, J, Schlegel, F, Cortes, J, Lavallee, T, Mehta, K, Young, A, Doti, L, Battaile, K.P, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Apo structure of (N1G37) Methyltransferase from Mycobacterium avium.
To Be Published
|
|
7RT0
 
 | |
3L5A
 
 | | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus | | Descriptor: | NADH/flavin oxidoreductase/NADH oxidase, TRIETHYLENE GLYCOL | | Authors: | Lam, R, Gordon, R.D, Vodsedalek, J, Battaile, K.P, Grebemeskel, S, Lam, K, Romanov, V, Chan, T, Mihajlovic, V, Thompson, C.M, Guthrie, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-21 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus
To be Published
|
|
3KWL
 
 | | Crystal structure of a hypothetical protein from Helicobacter pylori | | Descriptor: | uncharacterized protein | | Authors: | Lam, R, Thompson, C.M, Vodsedalek, J, Lam, K, Romanov, V, Battaile, K.P, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a hypothetical protein from Helicobacter pylori
To be Published
|
|
8EJX
 
 | |
4ZEG
 
 | | Crystal structure of TTK kinase domain in complex with a pyrazolopyrimidine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dual specificity protein kinase TTK, ... | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2015-04-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of TTK kinase domain in complex with a pyrazolopyrimidine inhibitor.
To Be Published
|
|
3UR6
 
 | | 1.5A resolution structure of apo Norwalk Virus Protease | | Descriptor: | 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
3JVO
 
 | | Crystal structure of bacteriophage HK97 gp6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Gp6 | | Authors: | Lam, R, Tuite, A, Battaile, K.P, Edwards, A.M, Maxwell, K.L, Chirgadze, N.Y. | | Deposit date: | 2009-09-17 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins.
J.Mol.Biol., 395, 2010
|
|
8G0G
 
 | | Crystal structure of diphtheria toxin H223Q/H257Q double mutant (pH 4.5) | | Descriptor: | ADENYLYL-3'-5'-PHOSPHO-URIDINE-3'-MONOPHOSPHATE, Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Ladokhin, A.S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine Protonation and Conformational Switching in Diphtheria Toxin Translocation Domain.
Toxins, 15, 2023
|
|
3K2A
 
 | | Crystal structure of the homeobox domain of human homeobox protein Meis2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Homeobox protein Meis2 | | Authors: | Lam, R, Soloveychik, M, Battaile, K.P, Romanov, V, Lam, K, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the homeobox domain of human homeobox protein Meis2
To be Published
|
|
8G0F
 
 | |
3MHZ
 
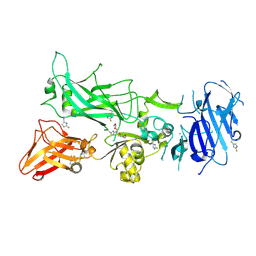 | | 1.7A structure of 2-fluorohistidine labeled Protective Antigen | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Wimalasena, D.S, Janowiak, B.E, Miyagi, M, Sun, J, Hajduch, J, Pooput, C, Kirk, K.L, Bann, J.G. | | Deposit date: | 2010-04-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence that histidine protonation of receptor-bound anthrax protective antigen is a trigger for pore formation.
Biochemistry, 49, 2010
|
|
3KOR
 
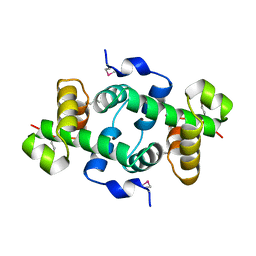 | | Crystal structure of a putative Trp repressor from Staphylococcus aureus | | Descriptor: | Possible Trp repressor | | Authors: | Lam, R, Vodsedalek, J, Lam, K, Romanov, V, Battaile, K.P, Beletskaya, I, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a putative Trp repressor from Staphylococcus aureus
To be Published
|
|
3UR9
 
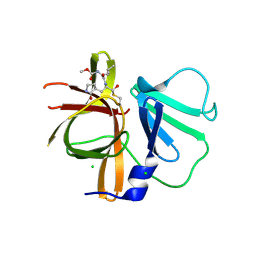 | | 1.65A resolution structure of Norwalk Virus Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like protease, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
8G1V
 
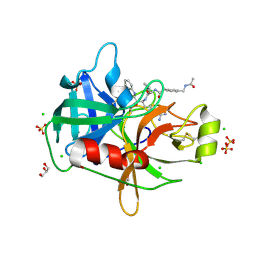 | | Crystal Structure Matriptase (C731S) in Complex with Inhibitor MM1132-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, N~2~-{[3-(acetamidomethyl)phenyl]acetyl}-N-[(2S)-1-(1,3-benzothiazol-2-yl)-5-carbamimidamido-1,1-dihydroxypentan-2-yl]-L-leucinamide, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Janetka, J.W. | | Deposit date: | 2023-02-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Use of protease substrate specificity screening in the rational design of selective protease inhibitors with unnatural amino acids: Application to HGFA, matriptase, and hepsin.
Protein Sci., 33, 2024
|
|
3LF5
 
 | | Structure of Human NADH cytochrome b5 oxidoreductase (Ncb5or) b5 Domain to 1.25A Resolution | | Descriptor: | Cytochrome b5 reductase 4, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Deng, B, Parthasarathy, S, Wang, W, Gibney, B.R, Battaile, K.P, Lovell, S, Benson, D.R, Zhu, H. | | Deposit date: | 2010-01-15 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Study of the individual cytochrome b5 and cytochrome b5 reductase domains of Ncb5or reveals a unique heme pocket and a possible role of the CS domain.
J.Biol.Chem., 285, 2010
|
|
3KBY
 
 | | Crystal structure of hypothetical protein from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein | | Authors: | Lam, R, Thompson, C.M, Battaile, K.P, Romanov, V, Kisselman, G, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-10-20 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein from Staphylococcus aureus
To be Published
|
|
3L20
 
 | | Crystal structure of a hypothetical protein from Staphylococcus aureus | | Descriptor: | Putative uncharacterized protein | | Authors: | Lam, R, Chan, T, Battaile, K.P, Mihajlovic, V, Romanov, V, Soloveychik, M, Kisselman, G, McGrath, T.E, Lam, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus
To be Published
|
|
5T6F
 
 | | 1.90 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (orthorhombic P form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
