1JJ2
 
 | | Fully Refined Crystal Structure of the Haloarcula marismortui Large Ribosomal Subunit at 2.4 Angstrom Resolution | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Klein, D.J, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The kink-turn: a new RNA secondary structure motif.
EMBO J., 20, 2001
|
|
7PMT
 
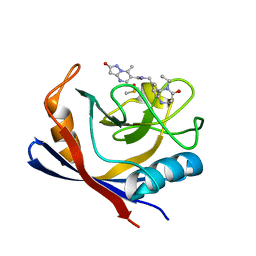 | | Human Cyclophilin D in complex with N-[(5-ethyl-4-oxo-1,2,3,4,5,6- hexahydro-1,5-benzodiazocin-8-yl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ... | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Human Cyclophilin D in complex with N-[(4-aminophenyl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
1KF6
 
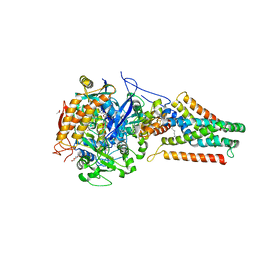 | | E. coli Quinol-Fumarate Reductase with Bound Inhibitor HQNO | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-19 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
7Q5C
 
 | | Crystal structure of OmpG in space group 96 | | Descriptor: | Outer membrane porin G, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Nguyen, T.T.M, Khan, A.R, Barringer, R, McManus, J.J, Race, P.R. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Experimental phase diagrams to optimise OmpG
To Be Published
|
|
1KVI
 
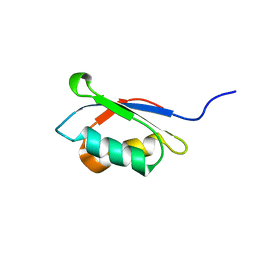 | |
1KVJ
 
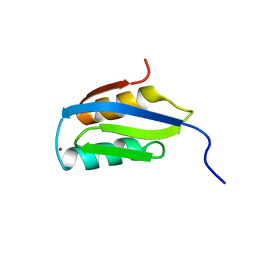 | |
1L16
 
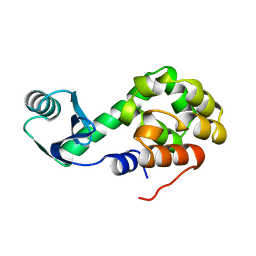 | |
1KFY
 
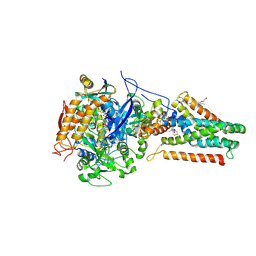 | | QUINOL-FUMARATE REDUCTASE WITH QUINOL INHIBITOR 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL | | Descriptor: | 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-24 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1LUP
 
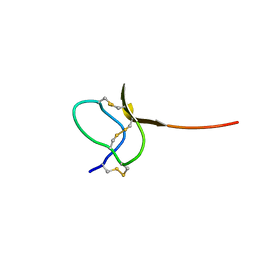 | | Solution structure of a toxin (GsMTx2) from the tarantula, Grammostola spatulata, which inhibits mechanosensitive ion channels | | Descriptor: | GsMTx2 | | Authors: | Oswald, R.E, Suchyna, T.M, McFeeters, R, Gottlieb, P, Sachs, F. | | Deposit date: | 2002-05-23 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of peptide toxins that block mechanosensitive ion channels
J.Biol.Chem., 277, 2002
|
|
1LWI
 
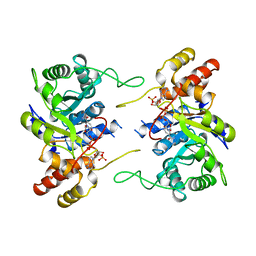 | | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE FROM RATTUS NORVEGICUS | | Descriptor: | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M.J, Schlegel, B.P, Jez, J.M, Penning, T.M, Lewis, M. | | Deposit date: | 1996-02-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
Biochemistry, 35, 1996
|
|
8OJR
 
 | | Solution NMR Structure of Alpha-Synuclein 1-25 Peptide in 50% TFE. | | Descriptor: | Alpha-synuclein | | Authors: | Allen, S.G, Williams, C, Meade, R.M, Tang, T.M.S, Crump, M.P, Mason, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-08-09 | | Method: | SOLUTION NMR | | Cite: | An N-terminal alpha-Synuclein fragment binds lipid vesicles to modulate lipid induced aggregation
Cell Rep Phys Sci, 2023
|
|
6HL5
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and ASPP1(932-954) | | Descriptor: | Apoptosis-stimulating of p53 protein 1, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Leissing, T.M, Chowdhury, R, Clifton, I.J, Lu, X, Schofield, C.J. | | Deposit date: | 2018-09-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and ASPP1(932-954)
To Be Published
|
|
5KL8
 
 | | Crystal structure of the Pumilio-Nos-CyclinB RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*UP*AP*UP*UP*UP*GP*UP*AP*AP*UP*U)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
5KMG
 
 | | Near-atomic cryo-EM structure of PRC1 bound to the microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kellogg, E.H, Howes, S, Ti, S.-C, Ramirez-Aportela, E, Kapoor, T.M, Chacon, P, Nogales, E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic cryo-EM structure of PRC1 bound to the microtubule.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KL0
 
 | | Crystal Structure of Phosphoglucomutase from Xanthomonas citri citri complexed with Glucose-1,6-biphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Goto, L.S, Pereira, H.M, Novo Mansur, M.T.M. | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and functional characterization of the phosphoglucomutase from Xanthomonas citri subsp. citri.
Biochim.Biophys.Acta, 1864, 2016
|
|
5KDO
 
 | | Heterotrimeric complex of the 4 alanine insertion variant of the Gi alpha1 subunit and the Gbeta1-Ggamma1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Conserved Hydrophobic Core in G alpha i1 Regulates G Protein Activation and Release from Activated Receptor.
J.Biol.Chem., 291, 2016
|
|
5KDL
 
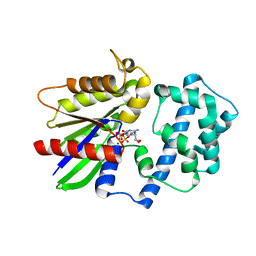 | | Crystal structure of the 4 alanine insertion variant of the Gi alpha1 subunit bound to GTPgammaS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | A Conserved Hydrophobic Core in G alpha i1 Regulates G Protein Activation and Release from Activated Receptor.
J.Biol.Chem., 291, 2016
|
|
5KDK
 
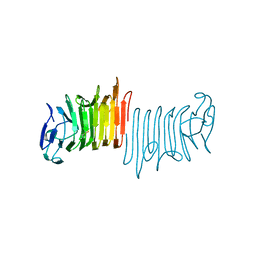 | |
5KLA
 
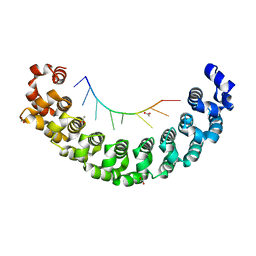 | |
5KEH
 
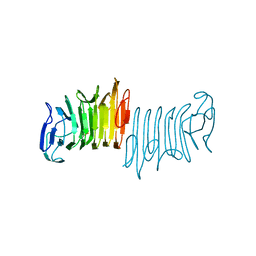 | |
5L08
 
 | | Cryo-EM structure of Casp-8 tDED filament | | Descriptor: | Caspase-8 | | Authors: | Fu, T.M, Li, Y, Lu, A, Wu, H. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Mol.Cell, 64, 2016
|
|
5KF3
 
 | |
5KIQ
 
 | | SrpA with sialyl LewisX | | Descriptor: | ACETATE ION, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | Structures of the Streptococcus sanguinis SrpA Binding Region with Human Sialoglycans Suggest Features of the Physiological Ligand.
Biochemistry, 2016
|
|
5KKD
 
 | |
6I6X
 
 | | New Irreversible a-l-Iduronidase Inhibitors and Activity-Based Probes | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},5~{S},6~{R})-7-methyl-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | New Irreversible alpha-l-Iduronidase Inhibitors and Activity-Based Probes.
Chemistry, 24, 2018
|
|
