1P5U
 
 | | X-ray structure of the ternary Caf1M:Caf1:Caf1 chaperone:subunit:subunit complex | | Descriptor: | Chaperone protein Caf1M, F1 capsule antigen | | Authors: | Zavialov, A.V, Berglund, J, Pudney, A.F, Fooks, L.J, Ibrahim, T.M, MacIntyre, S, Knight, S.D. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and Biogenesis of the Capsular F1 Antigen from Yersinia pestis. Preserved Folding Energy Drives Fiber Formation
Cell(Cambridge,Mass.), 113, 2003
|
|
5OPC
 
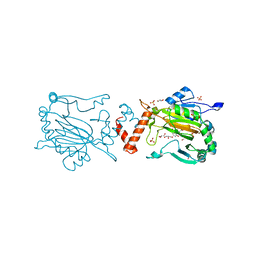 | | Factor Inhibiting HIF (FIH) in complex with zinc and Vadadustat | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Lu, X, Zhang, D. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
4UMQ
 
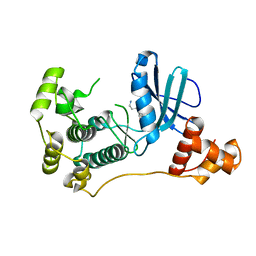 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-{5-[(3-hydroxy-5-methoxyphenyl)amino]-2-(phenylcarbamoyl)phenoxy}propan-1-aminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMT
 
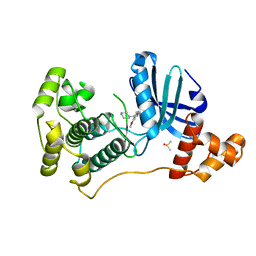 | | Structure of MELK in complex with inhibitors | | Descriptor: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMU
 
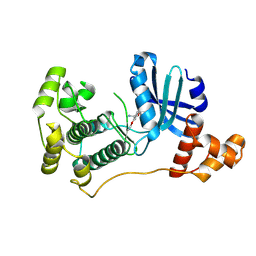 | | Structure of MELK in complex with inhibitors | | Descriptor: | (2-ethoxy-4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}phenyl)methanaminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMP
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-(isoquinolin-7-yl)prop-2-yn-1-ol, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMR
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 4-fluoro-N-(1,2,3,4-tetrahydroisoquinolin-7-yl)benzamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
1PDU
 
 | | Ligand-binding domain of Drosophila orphan nuclear receptor DHR38 | | Descriptor: | nuclear hormone receptor HR38 | | Authors: | Baker, K.D, Shewchuk, L.M, Korlova, T, Makishima, M, Hassell, A.M, Wisely, B, Caravella, J.A, Lambert, M.H, Wilson, T.M, Mangelsdorf, D.J. | | Deposit date: | 2003-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway.
Cell(Cambridge,Mass.), 113, 2003
|
|
5OVV
 
 | | PDZ domain from rat Shank3 bound to the C terminus of ProSAPiP1 | | Descriptor: | Leucine zipper putative tumor suppressor 3, SH3 and multiple ankyrin repeat domains protein 3, THIOCYANATE ION | | Authors: | Ponna, S.K, Myllykoski, M, Boeckers, T.M, Kursula, P. | | Deposit date: | 2017-08-29 | | Release date: | 2018-03-07 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.
J. Neurochem., 145, 2018
|
|
4V5Q
 
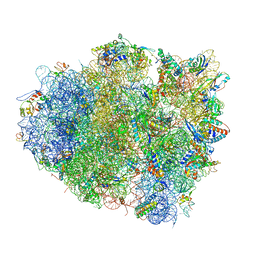 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4W8S
 
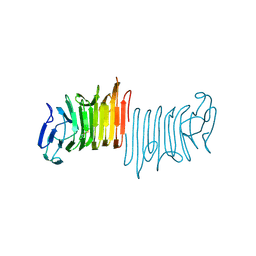 | |
1I8N
 
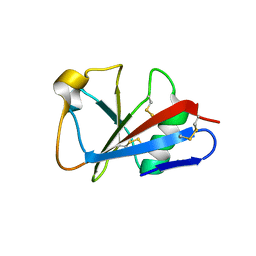 | | CRYSTAL STRUCTURE OF LEECH ANTI-PLATELET PROTEIN | | Descriptor: | ANTI-PLATELET PROTEIN, PROPIONAMIDE | | Authors: | Huizinga, E.G, Schouten, A, Connolly, T.M, Kroon, J, Sixma, J.J, Gros, P. | | Deposit date: | 2001-03-15 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of leech anti-platelet protein, an inhibitor of haemostasis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1P8D
 
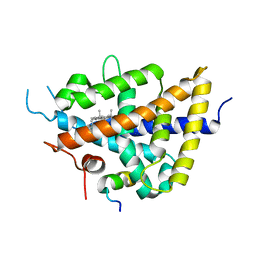 | | X-Ray Crystal Structure of LXR Ligand Binding Domain with 24(S),25-epoxycholesterol | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Oxysterols receptor LXR-beta, nuclear receptor coactivator 1 isoform 3 | | Authors: | Williams, S, Bledsoe, R.K, Collins, J.L, Boggs, S, Lambert, M.H, Miller, A.B, Moore, J, McKee, D.D, Moore, L, Nichols, J, Parks, D, Watson, M, Wisely, B, Willson, T.M. | | Deposit date: | 2003-05-06 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the liver X receptor beta ligand binding domain: regulation by
a histidine-tryptophan switch.
J.Biol.Chem., 278, 2003
|
|
1ONE
 
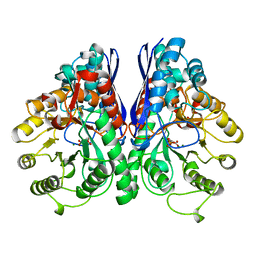 | | YEAST ENOLASE COMPLEXED WITH AN EQUILIBRIUM MIXTURE OF 2'-PHOSPHOGLYCEATE AND PHOSPHOENOLPYRUVATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, ENOLASE, MAGNESIUM ION, ... | | Authors: | Larsen, T.M, Wedekind, J.E, Rayment, I, Reed, G.H. | | Deposit date: | 1995-12-05 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution.
Biochemistry, 35, 1996
|
|
5WI7
 
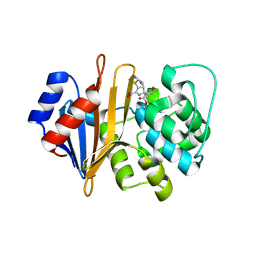 | | Structure of Acinetobacter baumannii carbapenemase OXA-239 K82D bound to doripenem | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, OXA-239 | | Authors: | Harper, T.M, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2017-07-18 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Multiple substitutions lead to increased loop flexibility and expanded specificity in Acinetobacter baumannii carbapenemase OXA-239.
Biochem. J., 475, 2018
|
|
5WNO
 
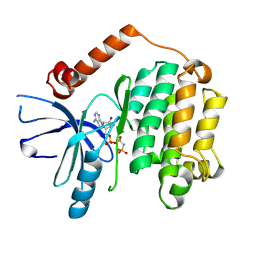 | | Crystal structure of C. elegans LET-23 kinase domain complexed with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase let-23 | | Authors: | Liu, L, Thaker, T.M, Jura, N. | | Deposit date: | 2017-08-01 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Regulation of Kinase Activity in the Caenorhabditis elegans EGF Receptor, LET-23.
Structure, 26, 2018
|
|
1OD8
 
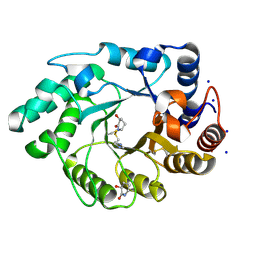 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine lactam | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, SODIUM ION, ... | | Authors: | Gloster, T.M, Roberts, S, Davies, G.J. | | Deposit date: | 2003-02-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A Xylobiose-Derived Isofagomine Lactam Glycosidase Inhibitor Binds as its Amide Tautomer
Chem.Commun.(Camb.), 8, 2003
|
|
5WIB
 
 | | Structure of Acinetobacter baumannii carbapenemase OXA-239 K82D bound to imipenem | | Descriptor: | Imipenem, OXA-239 | | Authors: | Harper, T.M, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2017-07-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Multiple substitutions lead to increased loop flexibility and expanded specificity in Acinetobacter baumannii carbapenemase OXA-239.
Biochem. J., 475, 2018
|
|
1OSV
 
 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
5WMX
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.M. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
1L0V
 
 | | Quinol-Fumarate Reductase with Menaquinol Molecules | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2002-02-13 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1PKN
 
 | | STRUCTURE OF RABBIT MUSCLE PYRUVATE KINASE COMPLEXED WITH MN2+, K+, AND PYRUVATE | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, PYRUVATE KINASE, ... | | Authors: | Larsen, T.M, Laughlin, L.T, Holden, H.M, Rayment, I, Reed, G.H. | | Deposit date: | 1994-03-25 | | Release date: | 1995-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of rabbit muscle pyruvate kinase complexed with Mn2+, K+, and pyruvate.
Biochemistry, 33, 1994
|
|
1NZK
 
 | | Crystal Structure of a Multiple Mutant (L44F, L73V, V109L, L111I, C117V) of Human Acidic Fibroblast Growth Factor | | Descriptor: | Acidic Fibroblast Growth Factor, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-02-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold.
Protein Sci., 12, 2003
|
|
5XV5
 
 | | Crystal structure of Rib7 mutant S88E from Methanosarcina mazei | | Descriptor: | Conserved protein | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
1P63
 
 | | Human Acidic Fibroblast Growth Factor. 140 Amino Acid Form with Amino Terminal His Tag and Leu111 Replaced with Ile (L111I) | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-04-28 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein
superfold
Protein Sci., 12, 2003
|
|
