5GK4
 
 | | Native structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 2.0 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 2.0 Angstrom resolution
To Be Published
|
|
5GK5
 
 | | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.9 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.9 angstrom resolution
To Be Published
|
|
5GK3
 
 | | Native structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution
To Be Published
|
|
5GK6
 
 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Citrate bound form | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Citrate bound form
To Be Published
|
|
5GK8
 
 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Acetate bound form | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Acetate bound form
To Be Published
|
|
5GK7
 
 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase bound to Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Tris bound form
To Be Published
|
|
1E50
 
 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
3VOP
 
 | | Structure of Vaccinia virus A27 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein A27 | | Authors: | Chang, T.H, Ko, T.P, Hsieh, F.L, Wang, A.H.J. | | Deposit date: | 2012-01-31 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of vaccinia viral A27 protein reveals a novel structure critical for its function and complex formation with A26 protein.
Plos Pathog., 9, 2013
|
|
4GH8
 
 | | Crystal structure of a 'humanized' E. coli dihydrofolate reductase | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | French, J.B, Liu, C.T, Hanoian, P, Pringle, T.H, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional significance of evolving protein sequence in dihydrofolate reductase from bacteria to humans.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7LXE
 
 | |
3LK0
 
 | | X-ray structure of bovine SC0067,Ca(2+)-S100B | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
4UE0
 
 | |
6XGU
 
 | | Crystal Structure of KRAS-Q61R (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | GLYCEROL, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6XI7
 
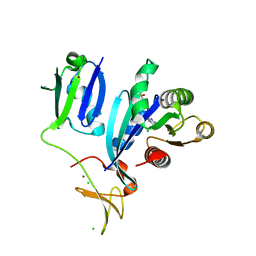 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form I) | | Descriptor: | CHLORIDE ION, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Chan, A.H, Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
3MFK
 
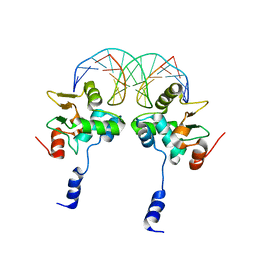 | |
4PHY
 
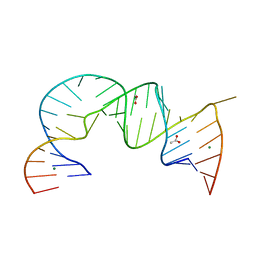 | | Functional conservation despite structural divergence in ligand-responsive RNA switches | | Descriptor: | ACETATE ION, MAGNESIUM ION, RNA (26-MER), ... | | Authors: | Boerneke, M.A, Dibrov, S.M, Hermann, T.H. | | Deposit date: | 2014-05-07 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional conservation despite structural divergence in ligand-responsive RNA switches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PTX
 
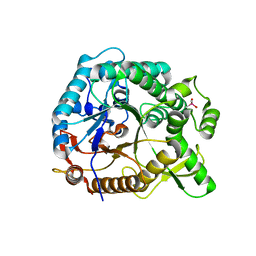 | | Halothermothrix orenii beta-glucosidase A, glucose complex | | Descriptor: | CACODYLATE ION, Glycoside hydrolase family 1, beta-D-glucopyranose | | Authors: | Hassan, N, Nguyen, T.H, Kori, L.D, Patel, B.K.C, Haltrich, D, Divne, C, Tan, T.C. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | Descriptor: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | Authors: | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2014-09-04 | | Release date: | 2015-07-01 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
5M9F
 
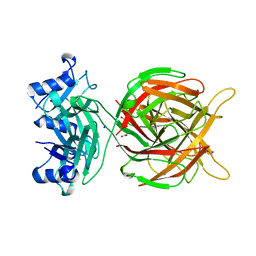 | |
6YRH
 
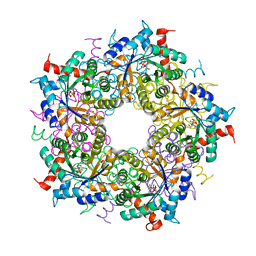 | |
6YRE
 
 | | Transaldolase variant T30C/D211C from T. acidophilum | | Descriptor: | ACETATE ION, GLYCEROL, Probable transaldolase | | Authors: | Sautner, V, Lietzow, T.H, Tittmann, K. | | Deposit date: | 2020-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Large-scale motions underlie physical but not chemical steps in transaldolase mechanism: Substrate binding by conformational selection and rate-determining product release
To Be Published
|
|
5N88
 
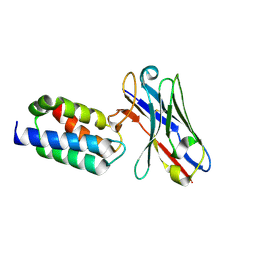 | | Crystal structure of antibody bound to viral protein | | Descriptor: | PC4 and SFRS1-interacting protein, VH59 antibody | | Authors: | Bao, L, Hannon, C, Cruz-Migoni, A, Ptchelkine, D, Sun, M.-y, Derveni, M, Bunjobpol, W, Chambers, J.S, Simmons, A, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein.
Sci Rep, 7, 2017
|
|
5AMO
 
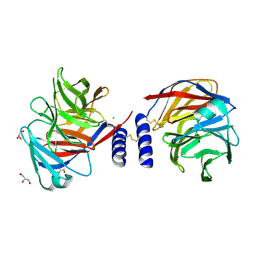 | | Structure of a mouse Olfactomedin-1 disulfide-linked dimer of the Olfactomedin domain and part of the coiled coil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Pronker, M.F, Bos, T.G.A.A, Sharp, T.H, Thies-Weesie, D.M, Janssen, B.J.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-04-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Olfactomedin-1 Has a V-Shaped Disulfide-Linked Tetrameric Structure.
J.Biol.Chem., 290, 2015
|
|
5DTJ
 
 | |
3D66
 
 | |
