6L7L
 
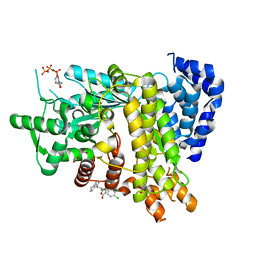 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide | | Descriptor: | 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
6L3R
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide | | Descriptor: | 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
2E4M
 
 | | Crystal structure of hemagglutinin subcomponent complex (HA-33/HA-17) from Clostridium botulinum serotype D strain 4947 | | Descriptor: | HA-17, Main hemagglutinin component | | Authors: | Hasegawa, K, Watanabe, T, Suzuki, T, Yamano, A, Niwa, K, Ohyama, T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Subunit Structure of Clostridium botulinum Serotype D Toxin Complex with Three Extended Arms
J.Biol.Chem., 282, 2007
|
|
5CI9
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 6 | | Descriptor: | 1-(propan-2-yl)-1H-benzimidazole-5-carboxylic acid, Protein Tob1, SODIUM ION | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
5CI8
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 1 | | Descriptor: | Protein Tob1, pyrrolo[1,2-a]quinoxalin-4(5H)-one | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
2D1P
 
 | | crystal structure of heterohexameric TusBCD proteins, which are crucial for the tRNA modification | | Descriptor: | Hypothetical UPF0116 protein yheM, Hypothetical UPF0163 protein yheN, Hypothetical protein yheL, ... | | Authors: | Numata, T, Fukai, S, Ikeuchi, Y, Suzuki, T, Nureki, O. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Sulfur Relay to RNA Mediated by Heterohexameric TusBCD Complex
Structure, 14, 2006
|
|
2D5U
 
 | |
2DER
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the initial tRNA binding state | | Descriptor: | PHOSPHATE ION, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DET
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the pre-reaction state | | Descriptor: | SULFATE ION, tRNA, tRNA-specific 2-thiouridylase mnmA | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DEU
 
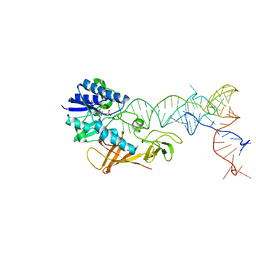 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the adenylated intermediate state | | Descriptor: | ADENOSINE MONOPHOSPHATE, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2UXD
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
2UXB
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
2UXC
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACGA, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
5EO7
 
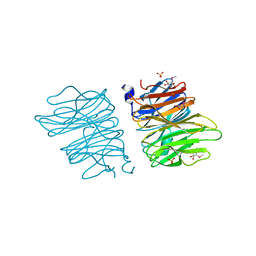 | | Crystal structure of AOL | | Descriptor: | Predicted protein, SULFATE ION, methyl 1-seleno-alpha-L-fucopyranoside | | Authors: | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5EO8
 
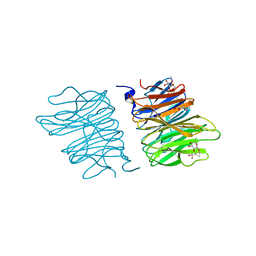 | | Crystal structure of AOL(868) | | Descriptor: | Predicted protein, methyl 1-seleno-beta-L-fucopyranoside | | Authors: | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5Y5N
 
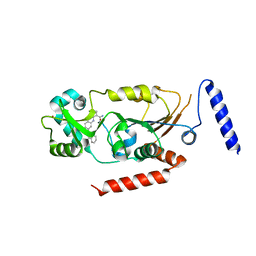 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
5B2H
 
 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
7E5O
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
5AXM
 
 | | Crystal structure of Thg1 like protein (TLP) with tRNA(Phe) | | Descriptor: | MAGNESIUM ION, RNA (75-MER), tRNA(His)-5'-guanylyltransferase (Thg1) like protein | | Authors: | Kimura, S, Suzuki, T, Yu, J, Kato, K, Yao, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Template-dependent nucleotide addition in the reverse (3'-5') direction by Thg1-like protein
Sci Adv, 2, 2016
|
|
