6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
2FM5
 
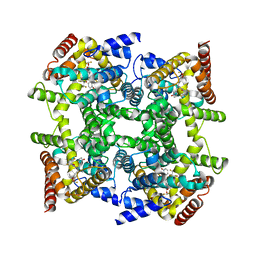 | | Crystal structure of PDE4D2 in complex with inhibitor L-869299 | | Descriptor: | (R)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHYL)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-07 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
2FM0
 
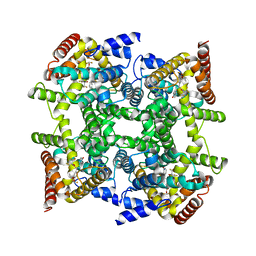 | | Crystal structure of PDE4D in complex with L-869298 | | Descriptor: | (S)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHY L)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
2N1K
 
 | |
7F0D
 
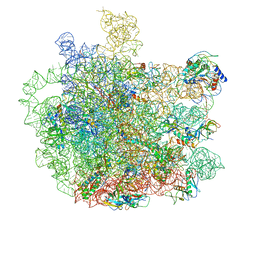 | | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosome subunit bound with clarithromycin | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Zhang, W, Sun, Y, Gao, N, Li, Z. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosomal subunit bound with clarithromycin reveals dynamic and specific interactions with macrolides.
Emerg Microbes Infect, 11, 2022
|
|
7U8G
 
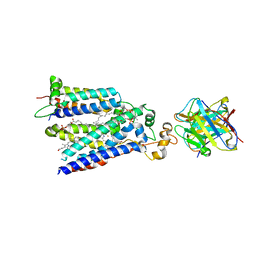 | | Cryo-EM structure of the core human NADPH oxidase NOX2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7G5 - heavy chain, ... | | Authors: | Noreng, S, Ota, N, Sun, Y, Masureel, M, Payandeh, J, Yi, T, Koerber, J.T. | | Deposit date: | 2022-03-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the core human NADPH oxidase NOX2.
Nat Commun, 13, 2022
|
|
7FIR
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with 1,4-mannobiose | | Descriptor: | Beta-1,2-mannobiose phosphorylase, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIP
 
 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIS
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with mannose 1-phosphate (M1P) | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, Beta-1,2-mannobiose phosphorylase, GLYCEROL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
2N79
 
 | | The structural and functional effects of the Familial Hypertrophic Cardiomyopathy-linked cardiac troponin C mutation, L29Q | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Sevrieva, I, Li, M.X, Irving, M, Sun, Y, Sykes, B. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural and functional effects of the familial hypertrophic cardiomyopathy-linked cardiac troponin C mutation, L29Q.
J.MOL.CELL.CARDIOL., 87, 2015
|
|
2L1R
 
 | | The structure of the calcium-sensitizer, dfbp-o, in complex with the N-domain of troponin C and the switch region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Robertson, I.M, Sun, Y, Li, M.X, Sykes, B.D. | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural and functional perspective into the mechanism of Ca(2+)-sensitizers that target the cardiac troponin complex.
J.MOL.CELL.CARDIOL., 49, 2010
|
|
2JXL
 
 | | Solution structure of cardiac N-domain troponin C mutant F77W-V82A | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Julien, O, Sun, Y, Wang, X, Lindhout, D.A, Thiessen, A, Irving, M, Sykes, B.D. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tryptophan Mutants of Cardiac Troponin C: 3D Structure, Troponin I Affinity, and in Situ Activity.
Biochemistry, 47, 2008
|
|
7C9V
 
 | | E30 F-particle in complex with FcRn | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, MYRISTIC ACID, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7DFV
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7C9S
 
 | | Echovirus 30 F-particle | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Wang, K, Sun, Y, Zhu, L, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7CRB
 
 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7C9U
 
 | | Echovirus 30 E-particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9T
 
 | | Echovirus 30 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
2KN0
 
 | | Solution NMR Structure of xenopus Fn14 | | Descriptor: | Fn14 | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-11 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2KMZ
 
 | | NMR Structure of hFn14 | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-10 | | Release date: | 2011-06-29 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2KN1
 
 | | Solution NMR Structure of BCMA | | Descriptor: | Tumor necrosis factor receptor superfamily member 17 | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-11 | | Release date: | 2011-02-23 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
