2P58
 
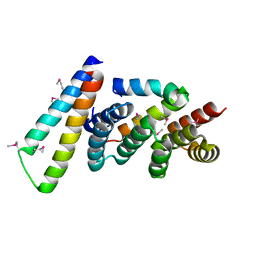 | | Structure of the Yersinia pestis Type III secretion system needle protein YscF in complex with its chaperones YscE/YscG | | Descriptor: | Putative type III secretion protein YscE, Putative type III secretion protein YscF, Putative type III secretion protein YscG | | Authors: | Sun, P, Austin, B.P, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG.
J.Mol.Biol., 377, 2008
|
|
2DL2
 
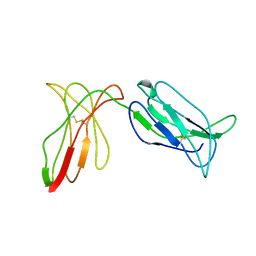 | | KILLER IMMUNOGLOBULIN RECEPTOR 2DL2 | | Descriptor: | PROTEIN (MHC CLASS I NK CELL RECEPTOR PRECURSOR (P58 NATURAL KILLER CELL RECEPTOR CLONE CL-43)) | | Authors: | Sun, P, Snyder, G. | | Deposit date: | 1999-03-08 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
9JPJ
 
 | |
9JPK
 
 | |
9JPL
 
 | |
6KE3
 
 | |
6KDF
 
 | | Crystal structure of the alpha beta heterodimer of human IDH3 in APO form. | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit beta | | Authors: | Sun, P, Ding, J. | | Deposit date: | 2019-07-02 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular basis for the function of the alpha beta heterodimer of human NAD-dependent isocitrate dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
6KDE
 
 | |
6L57
 
 | |
6L59
 
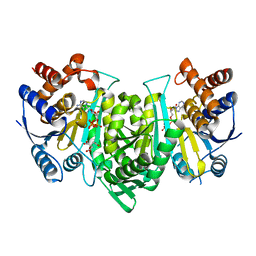 | | Crystal structure of the alpha gamma heterodimer of human IDH3 in complex with CIT, Mg and ATP binding at allosteric site and Mg, ATP binding at active site. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Isocitrate dehydrogenase [NAD] subunit alpha, ... | | Authors: | Sun, P, Ding, J. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Molecular mechanism of the dual regulatory roles of ATP on the alpha gamma heterodimer of human NAD-dependent isocitrate dehydrogenase.
Sci Rep, 10, 2020
|
|
8GS5
 
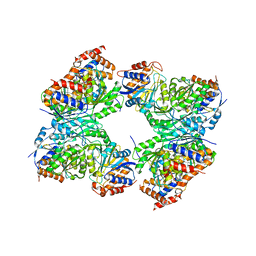 | | Crystal structure of a constitutively active mutant of human IDH3 holoenzyme in apo form | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.486 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRG
 
 | |
8GRH
 
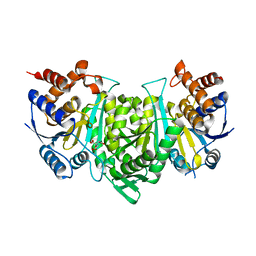 | | Crystal structure of a constitutively active mutant of the alpha gamma heterodimer of human IDH3 in complex with CIT | | Descriptor: | CITRIC ACID, Human IDH3 alpha subunit, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRU
 
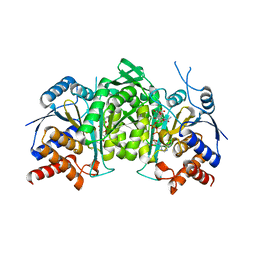 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ICT, NAD and Ca | | Descriptor: | CALCIUM ION, Human IDH3 alpha subunit, ISOCITRIC ACID, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRB
 
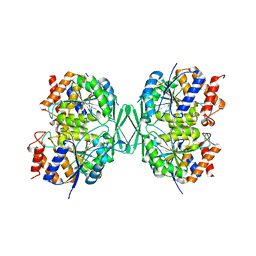 | |
6KDY
 
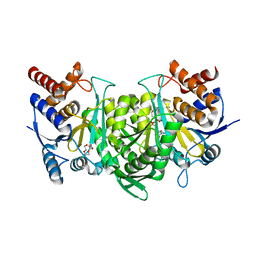 | |
2QWU
 
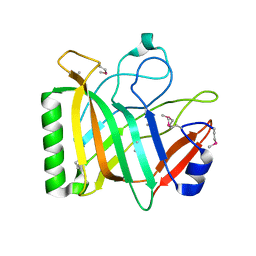 | |
1ZDE
 
 | | 1.95 Angstrom Crystal Structure of a dnaE Intein Precursor from Synechocystis Sp. Pcc 6803 | | Descriptor: | CALCIUM ION, DNA polymerase III alpha subunit, ZINC ION | | Authors: | Sun, P, Ye, S, Ferrandon, S, Evans, T.C, Xu, M.Q, Rao, Z. | | Deposit date: | 2005-04-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing
J.Mol.Biol., 353, 2005
|
|
1ZD7
 
 | | 1.7 Angstrom Crystal Structure Of Post-Splicing Form of a dnaE Intein from Synechocystis Sp. Pcc 6803 | | Descriptor: | DNA polymerase III alpha subunit, SULFATE ION | | Authors: | Sun, P, Ye, S, Ferrandon, S, Evans, T.C, Xu, M.Q, Rao, Z. | | Deposit date: | 2005-04-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing
J.Mol.Biol., 353, 2005
|
|
6CKF
 
 | | Structure of a new ShKT peptide from the sea anemone Oulactis sp: OspTx2a-p2 | | Descriptor: | OspTx2a-p2 | | Authors: | Sunanda, P, Krishnarjuna, B, Norton, R.S. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp.
Peptide Science, 110, 2018
|
|
6CKD
 
 | | Structure of a new ShKT peptide from the sea anemone Oulactis sp: OspTx2a-p1 | | Descriptor: | OspTx2a-p1 | | Authors: | Sunanda, P, Krishnarjuna, B, Norton, R.S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp.
Peptide Science, 110, 2018
|
|
1KCG
 
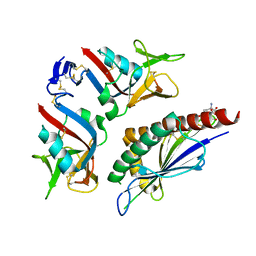 | | NKG2D in complex with ULBP3 | | Descriptor: | GLUTATHIONE, NKG2-D type II integral membrane protein, ULBP3 protein | | Authors: | Radaev, S, Sun, P. | | Deposit date: | 2001-11-08 | | Release date: | 2002-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3.
Immunity, 15, 2001
|
|
7ZE9
 
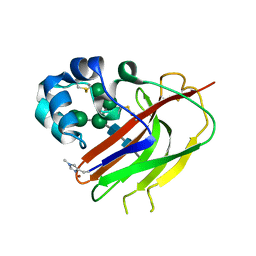 | | Structure of an AA16 LPMO-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Huang, Z, Banerjee, S, Muderspach, S.J, Sun, P, van Berkel, W.J.H, Kabel, M.A, Lo Leggio, L. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila.
Acs Catalysis, 13, 2023
|
|
8GRD
 
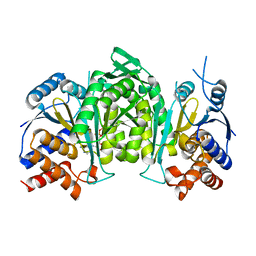 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, ... | | Authors: | Chen, X, Sun, P, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
